Transcription and Translation Dr.Osama AL-Mosawy
27 Slides600.50 KB

Transcription and Translation Dr.Osama AL-Mosawy
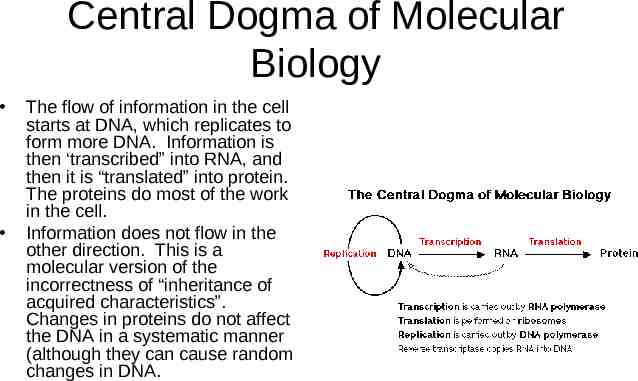
Central Dogma of Molecular Biology The flow of information in the cell starts at DNA, which replicates to form more DNA. Information is then ‘transcribed” into RNA, and then it is “translated” into protein. The proteins do most of the work in the cell. Information does not flow in the other direction. This is a molecular version of the incorrectness of “inheritance of acquired characteristics”. Changes in proteins do not affect the DNA in a systematic manner (although they can cause random changes in DNA.

RNA RNA plays a central role in the life of the cell. We are mostly going to look at its role in protein synthesis, but RNA does many other things as well. RNA can both store information (like DNA) and catalyze chemical reactions (like proteins). One theory for the origin of life has it starting out as RNA only, then adding DNA and proteins later. This theory is called the “RNA World”. RNA/protein hybrid structures are involved in protein synthesis (ribosome), splicing of messenger RNA, telomere maintenance, guiding ribosomes to the endoplasmic reticulum, and other tasks. Recently it has been found that very small RNA molecules are involves in gene regulation.
RNA Used in Protein Synthesis messenger RNA (mRNA). A copy of the gene that is being expressed. Groups of 3 bases in mRNA, called “codons” code for each individual amino acid in the protein made by that gene. – in eukaryotes, the initial RNA copy of the gene is called the “primary transcript”, which is modified to form mRNA. ribosomal RNA (rRNA). Four different RNA molecules that make up part of the structure of the ribosome. They perform the actual catalysis of adding an amino acid to a growing peptide chain. transfer RNA (tRNA). Small RNA molecules that act as adapters between the codons of messenger RNA and the amino acids they code for.

RNA vs. DNA RNA contains the sugar ribose; DNA contains deoxyribose. RNA contains the base uracil; DNA contains thymine instead. RNA is usually single stranded; DNA is usually double stranded. RNA is short: one gene long at most; DNA is long, containing many genes.
Transcription Transcription is the process of making an RNA copy of a single gene. Genes are specific regions of the DNA of a chromosome. The enzyme used in transcription is “RNA polymerase”. There are several forms of RNA polymerase. In eukaryotes, most genes are transcribed by RNA polymerase 2. The raw materials for the new RNA are the 4 ribonucleoside triphosphates: ATP, CTP, GTP, and UTP. It’s the same ATP as is used for energy in the cell. As with DNA replication, transcription proceeds 5- to 3’: new bases are added to the free 3’ OH group. Unlike replication, transcription does not need to build on a primer. Instead, transcription starts at a region of DNA called a “promoter”. For proteincoding genes, the promoter is located a few bases 5’ to (upstream from) the first base that is transcribed into RNA. Promoter sequences are very similar to each other, but not identical. If many promoters are compared, a “consensus sequence” can be derived. All promoters would be similar to this consensus sequence, but not necessarily identical.
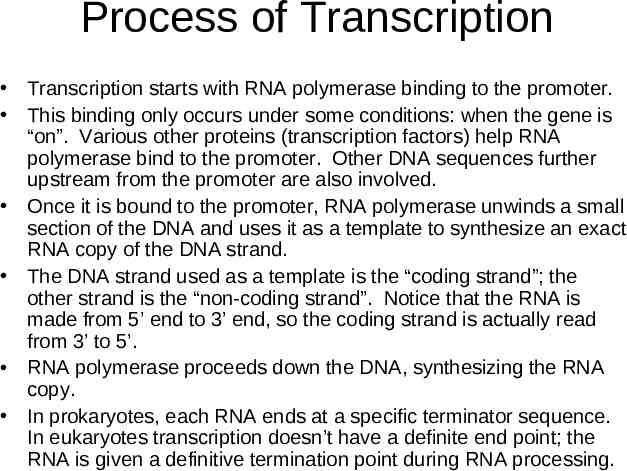
Process of Transcription Transcription starts with RNA polymerase binding to the promoter. This binding only occurs under some conditions: when the gene is “on”. Various other proteins (transcription factors) help RNA polymerase bind to the promoter. Other DNA sequences further upstream from the promoter are also involved. Once it is bound to the promoter, RNA polymerase unwinds a small section of the DNA and uses it as a template to synthesize an exact RNA copy of the DNA strand. The DNA strand used as a template is the “coding strand”; the other strand is the “non-coding strand”. Notice that the RNA is made from 5’ end to 3’ end, so the coding strand is actually read from 3’ to 5’. RNA polymerase proceeds down the DNA, synthesizing the RNA copy. In prokaryotes, each RNA ends at a specific terminator sequence. In eukaryotes transcription doesn’t have a definite end point; the RNA is given a definitive termination point during RNA processing.
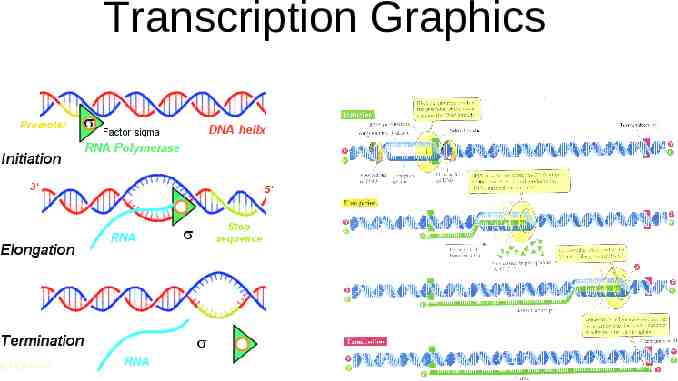
Transcription Graphics
After Transcription In prokaryotes, the RNA copy of a gene is messenger RNA, ready to be translated into protein. In fact, translation starts even before transcription is finished. In eukaryotes, the primary RNA transcript of a gene needs further processing before it can be translated. This step is called “RNA processing”. Also, it needs to be transported out of the nucleus into the cytoplasm. Steps in RNA processing: – 1. Add a cap to the 5’ end – 2. Add a poly-A tail to the 3’ end – 3. splice out introns.
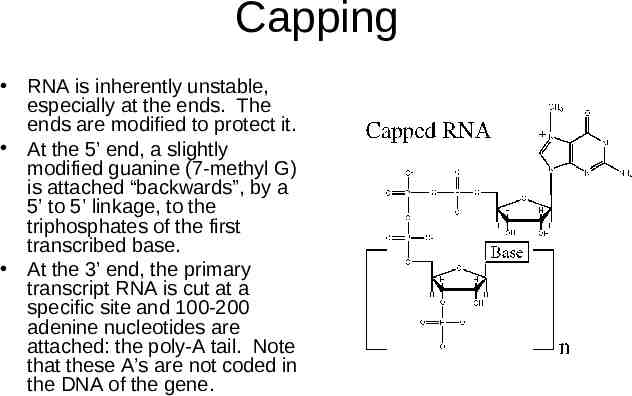
Capping RNA is inherently unstable, especially at the ends. The ends are modified to protect it. At the 5’ end, a slightly modified guanine (7-methyl G) is attached “backwards”, by a 5’ to 5’ linkage, to the triphosphates of the first transcribed base. At the 3’ end, the primary transcript RNA is cut at a specific site and 100-200 adenine nucleotides are attached: the poly-A tail. Note that these A’s are not coded in the DNA of the gene.
Introns Introns are regions within a gene that don’t code for protein and don’t appear in the final mRNA molecule. Protein-coding sections of a gene (called exons) are interrupted by introns. The function of introns remains unclear. They may help is RNA transport or in control of gene expression in some cases, and they may make it easier for sections of genes to be shuffled in evolution. But , no generally accepted reason for the existence of introns exists. There are a few prokaryotic examples, but most introns are found in eukaryotes. Some genes have many long introns: the dystrophin gene (mutants cause muscular dystrophy) has more than 70 introns that make up more than 99% of the gene’s sequence. However, not all eukaryotic genes have introns: histone genes, for example, lack introns.
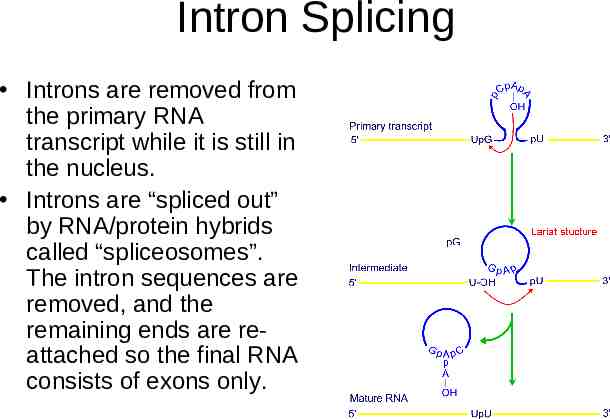
Intron Splicing Introns are removed from the primary RNA transcript while it is still in the nucleus. Introns are “spliced out” by RNA/protein hybrids called “spliceosomes”. The intron sequences are removed, and the remaining ends are reattached so the final RNA consists of exons only.
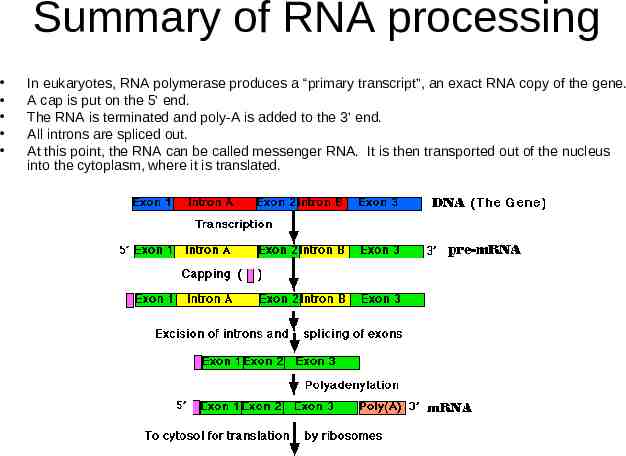
Summary of RNA processing In eukaryotes, RNA polymerase produces a “primary transcript”, an exact RNA copy of the gene. A cap is put on the 5’ end. The RNA is terminated and poly-A is added to the 3’ end. All introns are spliced out. At this point, the RNA can be called messenger RNA. It is then transported out of the nucleus into the cytoplasm, where it is translated.
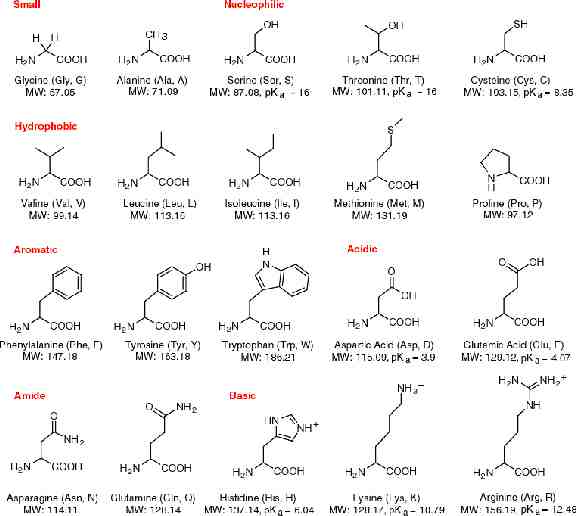
Proteins Proteins are composed of one or more polypeptides, plus (in some cases) additional small molecules (cofactors). Polypeptides are linear chains of amino acids. After synthesis, the new polypeptide folds spontaneously into its active configuration and combines with the other necessary subunits to form an active protein. Thus, all the information necessary to produce the protein is contained in the DNA base sequence that codes for the polypeptides. The sequence of amino acids in a polypeptide is known as its “primary structure”.
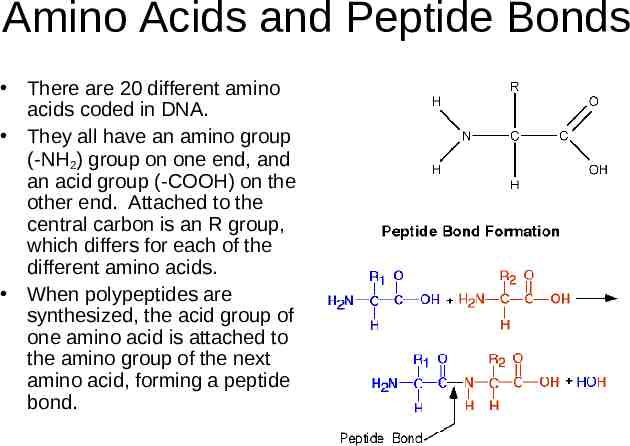
Amino Acids and Peptide Bonds There are 20 different amino acids coded in DNA. They all have an amino group (-NH2) group on one end, and an acid group (-COOH) on the other end. Attached to the central carbon is an R group, which differs for each of the different amino acids. When polypeptides are synthesized, the acid group of one amino acid is attached to the amino group of the next amino acid, forming a peptide bond.
Translation Translation of mRNA into protein is accomplished by the ribosome, an RNA/protein hybrid. Ribosomes are composed of 2 subunits, large and small. Ribosomes bind to the translation initiation sequence on the mRNA, then move down the RNA in a 5’ to 3’ direction, creating a new polypeptide. The first amino acid on the polypeptide has a free amino group, so it is called the “N-terminal”. The last amino acid in a polypeptide has a free acid group, so it is called the “Cterminal”. Each group of 3 nucleotides in the mRNA is a “codon”, which codes for 1 amino acids. Transfer RNA is the adapter between the 3 bases of the codon and the corresponding amino acid.

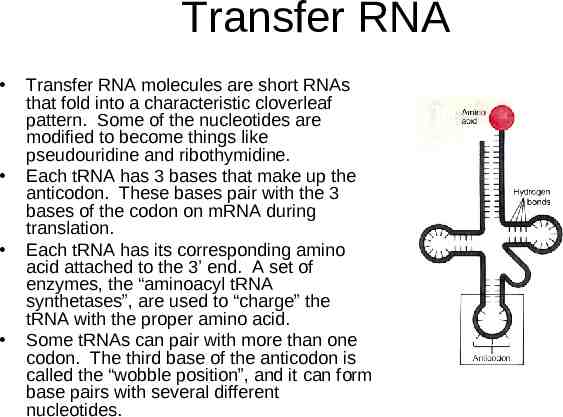
Transfer RNA Transfer RNA molecules are short RNAs that fold into a characteristic cloverleaf pattern. Some of the nucleotides are modified to become things like pseudouridine and ribothymidine. Each tRNA has 3 bases that make up the anticodon. These bases pair with the 3 bases of the codon on mRNA during translation. Each tRNA has its corresponding amino acid attached to the 3’ end. A set of enzymes, the “aminoacyl tRNA synthetases”, are used to “charge” the tRNA with the proper amino acid. Some tRNAs can pair with more than one codon. The third base of the anticodon is called the “wobble position”, and it can form base pairs with several different nucleotides.
Initiation of Translation In prokaryotes, ribosomes bind to specific translation initiation sites. There can be several different initiation sites on a messenger RNA: a prokaryotic mRNA can code for several different proteins. Translation begins at an AUG codon, or sometimes a GUG. The modified amino acid N-formyl methionine is always the first amino acid of the new polypeptide. In eukaryotes, ribosomes bind to the 5’ cap, then move down the mRNA until they reach the first AUG, the codon for methionine. Translation starts from this point. Eukaryotic mRNAs code for only a single gene. (Although there are a few exceptions, mainly among the eukaryotic viruses). Note that translation does not start at the first base of the mRNA. There is an untranslated region at the beginning of the mRNA, the 5’ untranslated region (5’ UTR).

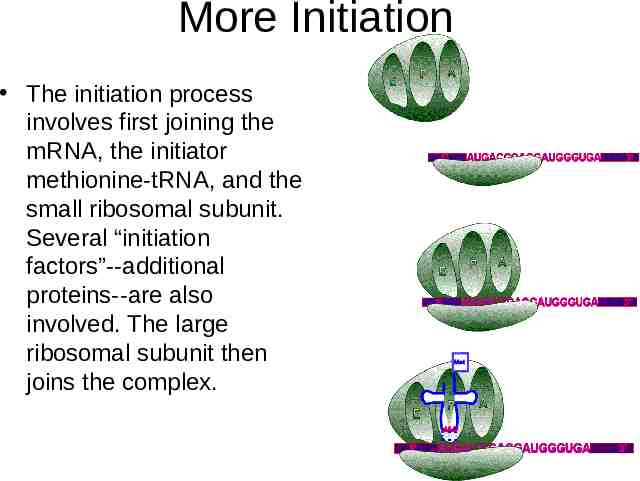
More Initiation The initiation process involves first joining the mRNA, the initiator methionine-tRNA, and the small ribosomal subunit. Several “initiation factors”--additional proteins--are also involved. The large ribosomal subunit then joins the complex.
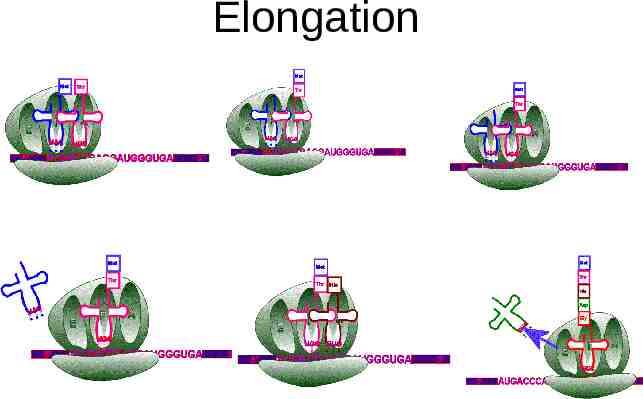
Elongation The ribosome has 2 sites for tRNAs, called P and A. The initial tRNA with attached amino acid is in the P site. A new tRNA, corresponding to the next codon on the mRNA, binds to the A site. The ribosome catalyzes a transfer of the amino acid from the P site onto the amino acid at the A site, forming a new peptide bond. The ribosome then moves down one codon. The now-empty tRNA at the P site is displaced off the ribosome, and the tRNA that has the growing peptide chain on it is moved from the A site to the P site. The process is then repeated: – the tRNA at the P site holds the peptide chain, and a new tRNA binds to the A site. – the peptide chain is transferred onto the amino acid attached to the A site tRNA. – the ribosome moves down one codon, displacing the empty P site tRNA and moving the tRNA with the peptide chain from the A site to the P site.

Elongation
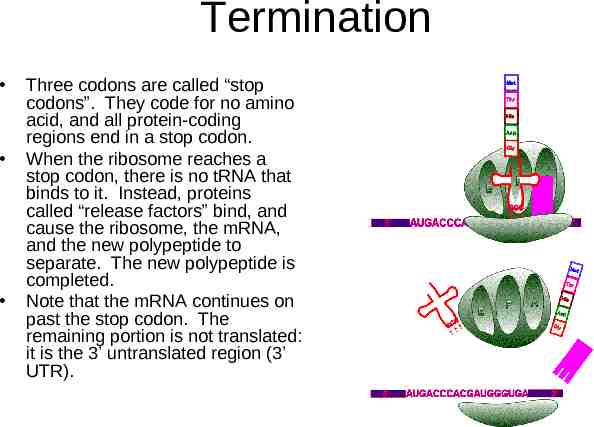
Termination Three codons are called “stop codons”. They code for no amino acid, and all protein-coding regions end in a stop codon. When the ribosome reaches a stop codon, there is no tRNA that binds to it. Instead, proteins called “release factors” bind, and cause the ribosome, the mRNA, and the new polypeptide to separate. The new polypeptide is completed. Note that the mRNA continues on past the stop codon. The remaining portion is not translated: it is the 3’ untranslated region (3’ UTR).
Post-Translational Modification New polypeptides usually fold themselves spontaneously into their active conformation. However, some proteins are helped and guided in the folding process by chaperone proteins Many proteins have sugars, phosphate groups, fatty acids, and other molecules covalently attached to certain amino acids. Most of this is done in the endoplasmic reticulum. Many proteins are targeted to specific organelles within the cell. Targeting is accomplished through “signal sequences” on the polypeptide. In the case of proteins that go into the endoplasmic reticulum, the signal seqeunce is a group of amino acids at the N terminal of the polypeptide, which are removed from the final protein after translation.
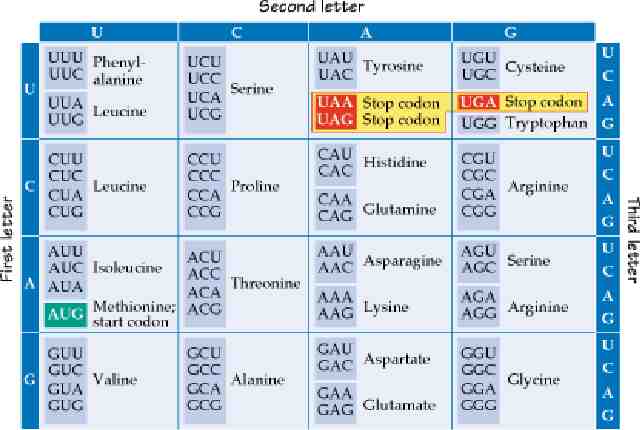
The Genetic Code Each group of 3 nucleotides on the mRNA is a codon. Since there are 4 bases, there are 43 64 possible codons, which must code for 20 different amino acids. More than one codon is used for most amino acids: the genetic code is “degenerate”. This means that it is not possible to take a protein sequence and deduce exactly the base sequence of the gene it came from. In most cases, the third base of the codon (the wobble base) can be altered without changing the amino acid. AUG is used as the start codon. All proteins are initially translated with methionine in the first position, although it is often removed after translation. There are also internal methionines in most proteins, coded by the same AUG codon. There are 3 stop codons, also called “nonsense” codons. Proteins end in a stop codon, which codes for no amino acid.
More Genetic Code The genetic code is almost universal. It is used in both prokaryotes and eukaryotes. However, some variants exist, mostly in mitochondria which have very few genes. For instance, CUA codes for leucine in the universal code, but in yeast mitochondria it codes for threonine. Similarly, AGA codes for arginine in the universal code, but in human and Drosophila mitochondria it is a stop codon. There are also a few known variants in the code used in nuclei, mostly among the protists.