BioNetFit Design Review 2 Team U.I. Fit Charles Chatwin, Matthew
23 Slides5.05 MB

BioNetFit Design Review 2 Team U.I. Fit Charles Chatwin, Matthew Burns, Joshua Gutman, Tanner Brelje Mentor & Client: Dr. Abolfazl Razi 1

Team U.I. Fit Charles Chatwin Matthew Burns Team Leader Web Designer Tanner Brelje Joshua Gutman Database Designer Release Manager Charles 2
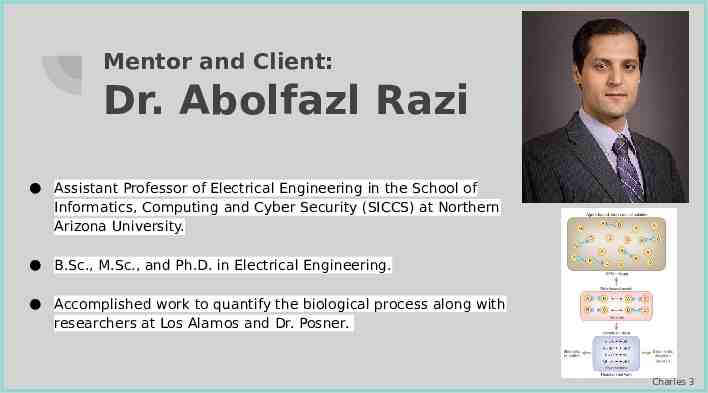
Mentor and Client: Dr. Abolfazl Razi Assistant Professor of Electrical Engineering in the School of Informatics, Computing and Cyber Security (SICCS) at Northern Arizona University. B.Sc., M.Sc., and Ph.D. in Electrical Engineering. Accomplished work to quantify the biological process along with researchers at Los Alamos and Dr. Posner. Charles 3
Molecular Biology Molecular Biology is an ever-advancing scientific field that finds a main focus in the experimentation and testing of molecular combinations. In order to make computations and develop headway in the field, researchers are required to test the interactions of molecules in a laboratory setting. These experiments consume a heavy amount of time and resources, and may not even yield palpable results. Charles 4
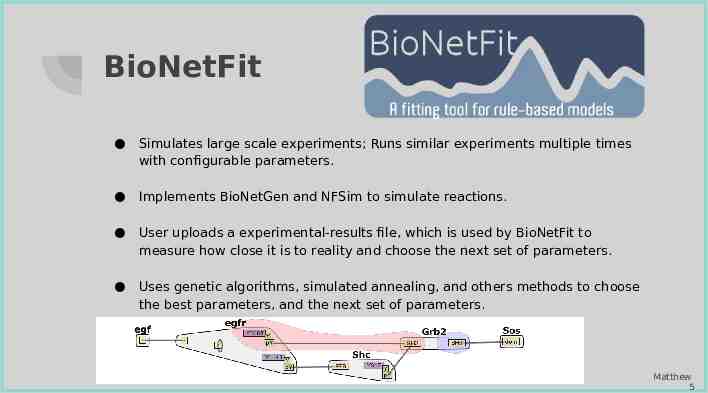
BioNetFit Simulates large scale experiments; Runs similar experiments multiple times with configurable parameters. Implements BioNetGen and NFSim to simulate reactions. User uploads a experimental-results file, which is used by BioNetFit to measure how close it is to reality and choose the next set of parameters. Uses genetic algorithms, simulated annealing, and others methods to choose the best parameters, and the next set of parameters. Matthew 5
The Problem BioNetFit suffers from poor usability. Only exists as a command-line tool. Results are not easily interpreted. No visual representation of the data. Cannot run large experiments in a reasonable timeframe. Works accurately, but very slowly on a single machine. Matthew 6
Image of bionetfit on cmd line Example output Example config file Matthew 7
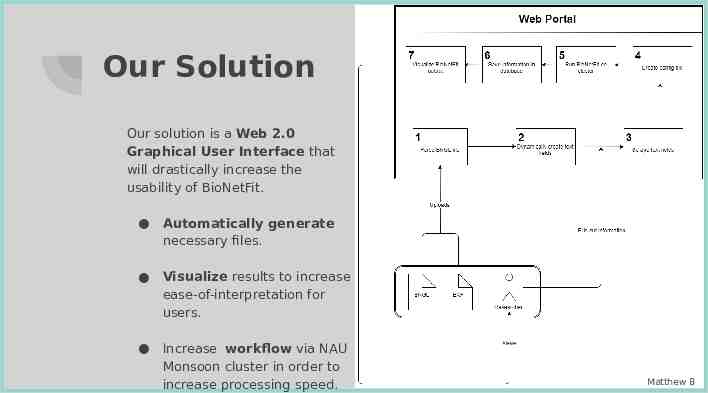
Our Solution Our solution is a Web 2.0 Graphical User Interface that will drastically increase the usability of BioNetFit. Automatically generate necessary files. Visualize results to increase ease-of-interpretation for users. Increase workflow via NAU Monsoon cluster in order to increase processing speed. Matthew 8
Key User Requirements The user must be able to: Generate configuration files. Visualize the results of BioNetFit Save all information related to BioNetFit in a database. Repeat past experiments with small alterations. Download any and all files related to BioNetFit. Matthew 9

Key Technical Requirements Dynamic HTML generation Scraping text fields A configuration file is created from user-inputted information Storing information in a database Text fields are scraped to get user-inputted information Creating files from user-inputted information Text fields are created from user-uploaded file All input and output files are stored in a database for each user Visualizing data Plot BioNetFit output Matthew 10
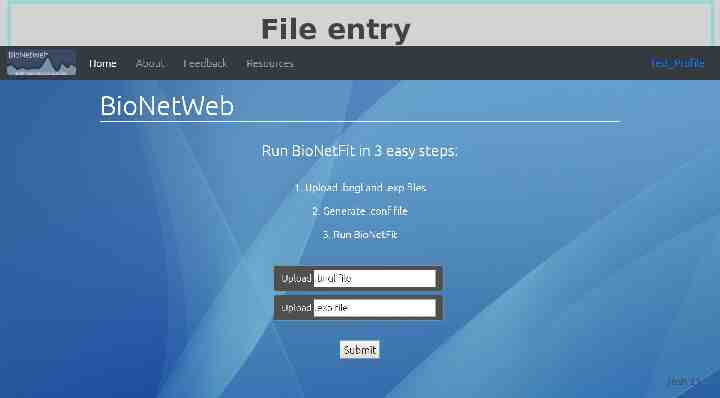
File entry Josh 11
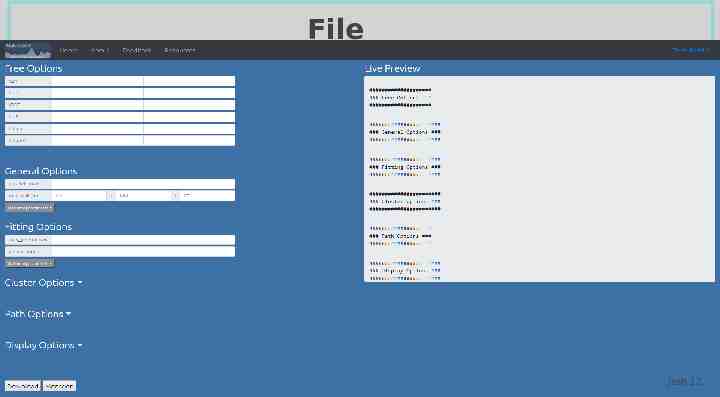
File Generation Josh 12
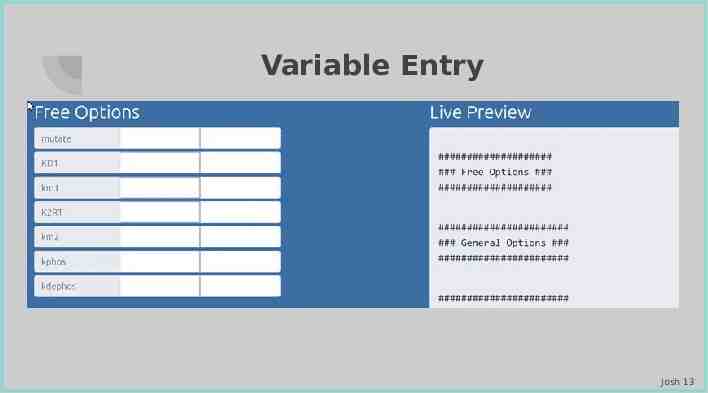
Variable Entry Josh 13
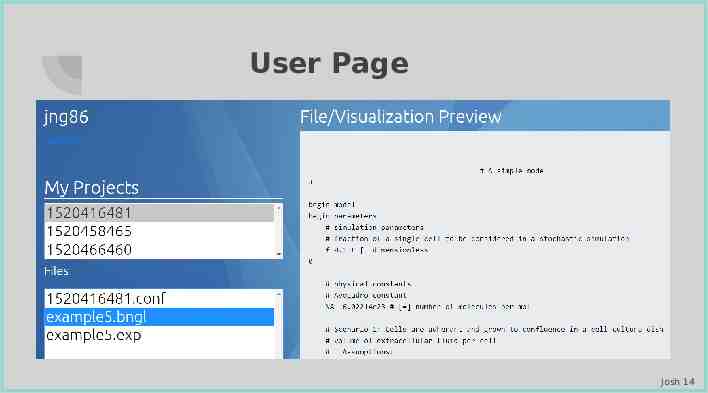
User Page Josh 14
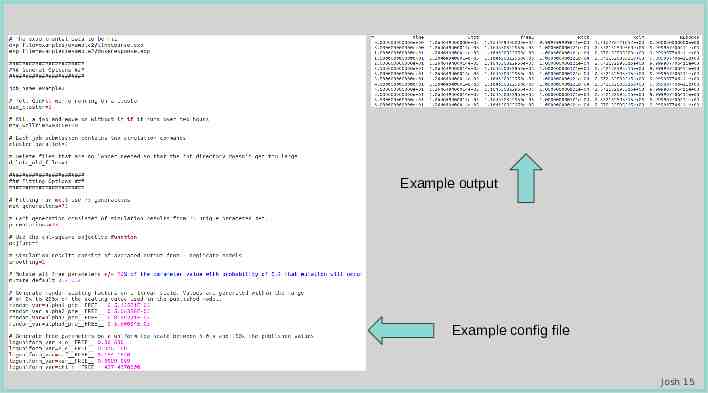
Image of bionetfit on cmd line Example output Example config file Josh 15
Josh 16

Python Framework Requirement Fulfilled by Python? Dynamic HTML generation Django Yes Scraping Text fields Django Yes Creating files Vanilla Python Yes SSH from server to server Paramiko Yes Database MongoDB Partial Visualizing Data D3 No Josh 17
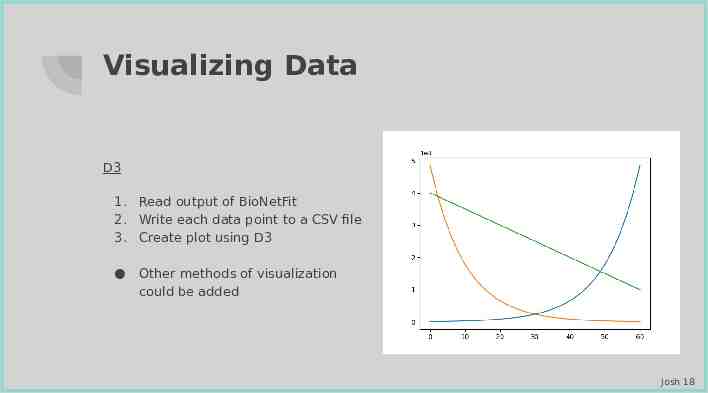
Visualizing Data D3 1. Read output of BioNetFit 2. Write each data point to a CSV file 3. Create plot using D3 Other methods of visualization could be added Josh 18
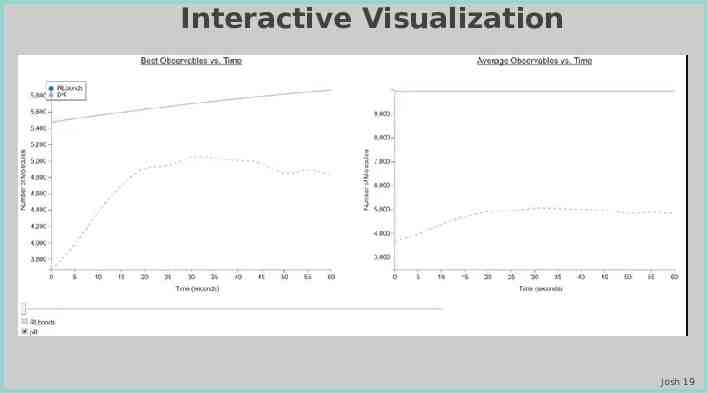
Interactive Visualization Josh 19
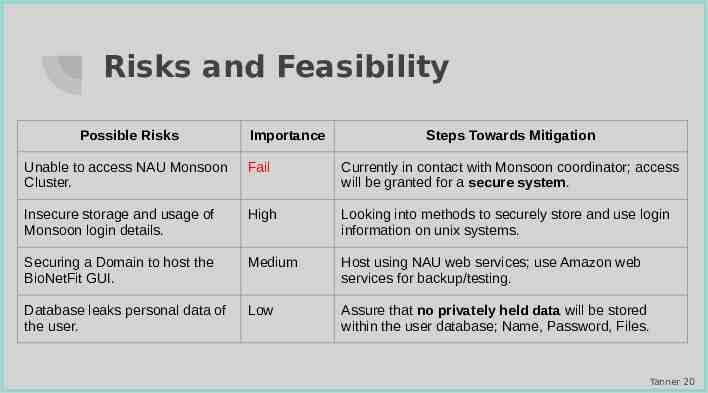
Risks and Feasibility Possible Risks Importance Steps Towards Mitigation Unable to access NAU Monsoon Cluster. Fail Currently in contact with Monsoon coordinator; access will be granted for a secure system. Insecure storage and usage of Monsoon login details. High Looking into methods to securely store and use login information on unix systems. Securing a Domain to host the BioNetFit GUI. Medium Host using NAU web services; use Amazon web services for backup/testing. Database leaks personal data of the user. Low Assure that no privately held data will be stored within the user database; Name, Password, Files. Tanner 20

Successes and Failures Successes: Database successfully implemented without use of personal data. Full pythonic implementation of website as well as additional features. Dynamic creation of files, upload and download functionality. Failures: Access to the Monsoon servers has been denied due to possible security risks. Preliminary visualization methods deemed inadequate. Tanner 21
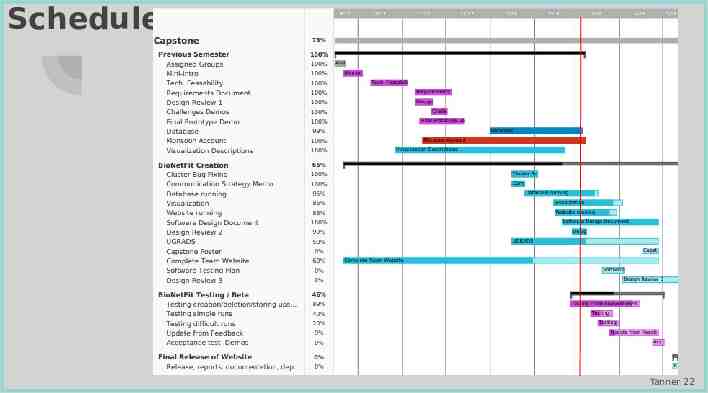
Schedule Tanner 22

Conclusion The Problem: BioNetFit is not easy to use, requires a lot of time to process huge experiments, and gives outputs that are hard for researchers to digest. Our Solution: A Web 2.0 GUI that will increase ease-of-use, implemented with parallelization on the Monsoon cluster, and clear visualizations of outputs. Implementation via Python Framework Django, Paramiko, vanilla Python, etc. Risks to design can be overcome via feasible options. Tanner 23