Chapter 1 Introduction, History of molecular Evolution Welcome to the
39 Slides2.95 MB

Chapter 1 Introduction, History of molecular Evolution Welcome to the 2011 class Chau-Ti Ting [email protected] Unless noted, the course materials are licensed under Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Taiwan (CC BY-NC-SA 3.0) 1
Course description: This course is to introduce the fundamentals and methodology on research of molecular evolution. The topics include evolutionary changes and patterns of sequences and methods for phylogenetic analyses. Topics on recent advances of molecular evolution studies will be offered at the end, on phylogenomics, horizontal gene transfer, and genome evolution. Students are highly recommended to take evolution, genetics or equivalent subjects before taking this course. There are three laboratory practices for students being familiar with data retrieving and analyses. Several homework assignments will be distributed during the semester, which require substantial extra work time after the class. 2

Lecture Wednesday 10:20 12:10, Life Science 419 Instructors Chau-Ti Ting ( 丁照棣 ) E-mail: ctting at ntu.edu.tw Phone: 3366 2522 Jer-Ming Hu ( 胡哲明 ) E-mail: jmhu at ntu.edu.tw Phone: 3366 2472 3
Course website: http://ceiba.ntu.edu.tw/1002molevo Official language: English Prerequisite: Biology; Evolutionary Biology; Genetics or equivalent preferred Office hour: Wed 12:00-13:00, or with appointments 4
Laboratory practice We will have computers available to use on the class, but you are welcome to bring your own laptop computer. In the later case, you should download the corresponding softwares before the class. You are welcome to use your own data for the homework assignments. 5

TA Grading Midterm exam 25% Final exam 30% Homework 35% Classroom performance 10% 6

Textbook There is no required textbook for this class. Most of the lectures are based on selected chapters from the following books. 7
Suggested reading Graur, D. and W.-H. Li. 2000. Fundamentals of Molecular Evolution. 2nd ed., Sinauer Assoc., Sunderland, MA, USA. Hall, B. G. 2011. Phylogenetic trees made easy: a how-to manual, 4th ed. Sinauer Assoc., Inc., Sunderland, MA, USA. Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA, USA. Lynch, M. 2007. The origins of genome architecture. Sinauer Associates, Sunderland, MA, USA. 8

Fundamentals of Molecular Evolution by Dan Graur and Wen-Hsiung Li, 2nd Edition (1999, Sinauer) http://www.sinauer.com/detail.php?id 2666 9

Hall, B. G. 2011. Phylogenetic trees made easy: a how-to manual, 4th ed. Sinauer Assoc., Inc., Sunderland, MA, USA. http://www.sinauer.com/detail.php?id 6069 10

Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA, USA. http://books.google.com.tw/books/about/Molecular evolution.ht ml?id 2mPGQgAACAAJ&redir esc y 11

Lynch, M. 2007. The origins of genome architecture. Sinauer Associates, Sunderland, MA, USA. http://www.sinauer.com/detail.php?id 4843 12
Molecular Evolution and Phylogenetics by Masatoshi Nei, Sudhir Kumar Oxford University Press (2000) http://www.oup.com/us/catalog/general/subject/LifeSciences/EvolutionaryBiology/?view usa&ci 97 5855 Molecular Evolution : A Phylogenetic Approach by Roderic D. M. Page, Edward C. Holmes Blackwell Science Inc. (1998) http://as.wiley.com/WileyCDA/WileyTitle/productCd-0865428891.html Integrated Molecular Evolution by Scott O. Rogers (Jul 27, 2011) http://www.crcpress.com/product/isbn/9781439819951;jsessionid 43hickpiklcWh5SL-z17 dQ** 13

More reference book Bioinformatics and Molecular Evolution by Paul Higgs and Teresa Attwood (Wiley-Blackwell, 2005) *Computational Molecular Evolution by Ziheng Yang (Oxford University Press, 2006) *Inferring Phylogenies (Paperback) by Joseph Felsenstein (Sinauer, 2003) *Statistical Methods in Molecular Evolution by Rasmus Nielsen (Springer, 2005) The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny Ed. by Marco Salemi and Anne-Mieke Vandamme (Cambridge University Press, 2003) 14

Journals Cladistics (http://www.blackwell-synergy.com/toc/cla/21/3;jsessionid bfrbfx Qdkn8) BMC Evol Biol (http://www.biomedcentral.com/bmcevolbiol/) Evolution (http://www.blackwell-synergy.com/loi/EVO) Evol Dev (http://www.blackwell-synergy.com/loi/ede) Evol Bioinformatics (http://la-press.com/journal.php?journal id 17&issue id 103) Genome Biology (http://genomebiology.com/home/) J Evol Biol (http://www.blackwellpublishing.com/jeb enhanced/) J Mol Evol (http://www.springer.com/life sci/cell biology/journal/239) Mol Biol Evol (http://mbe.oxfordjournals.org/) Mol Ecol (http://www.blackwell-synergy.com/loi/MEC) Mol Phyl Evol (http://www.sciencedirect.com/science/journal/10557903) Syst Biol (http://www.informaworld.com/smpp/title db bisc content .ueslist) Trends in Ecology & Evolution (http://www.sciencedirect.com/science/ )15

Related courses and workshops NTU campus Mathematical Molecular Evolution (221 U4730) Molecular Evolution Laboratory (B44 U1630) Population Genetics (B44 U1570) Seminar in Molecular Evolution (B41 M0310) Summer course on molecular phylogeny Workshop on Computational Molecular Evolution ( http://homepage.ntu.edu.tw/ ctting/CMB.html) Elsewhere Summer Institute in Statistical Genetics (http://www.biostat.washington.edu/node/1020) Workshop on Molecular Evolution ( http://workshop.molecularevolution.org/mbl/) 16
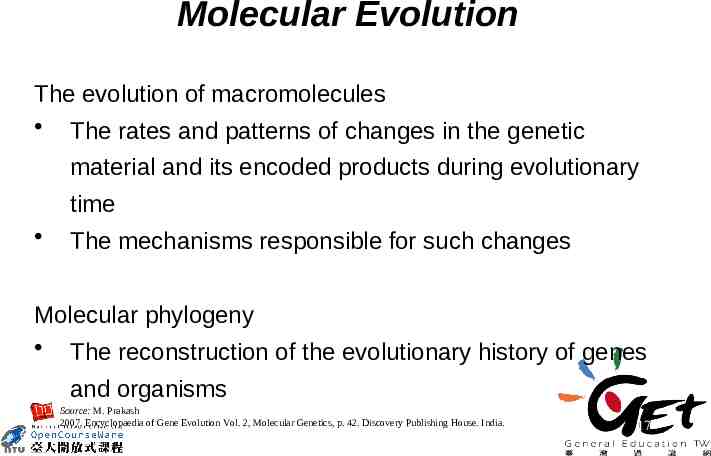
Molecular Evolution The evolution of macromolecules The rates and patterns of changes in the genetic material and its encoded products during evolutionary time The mechanisms responsible for such changes Molecular phylogeny The reconstruction of the evolutionary history of genes and organisms Source: M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 42. Discovery Publishing House. India. 17

Carolina Biological Supply Company Source: Carolina Biological Supply Company http://www.geneticorigins.org/ Science /AAAS Source: 2001. Science. Vol. 291. Issue 5507. http://www.sciencemag.org/content/291/5507.cover-expansion 18

1904 Nuttall conducted precipitin tests of serum proteins to infer the phylogenetic relationship among various groups of animals. Source: M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 42. Discovery Publishing House. India. George H. F. Nuttall 1904. Blood immunity and blood relationship. University press. Cambridge, England. http://www.biodiversitylibrary.org/title/8418#page/7/mode/1up 19
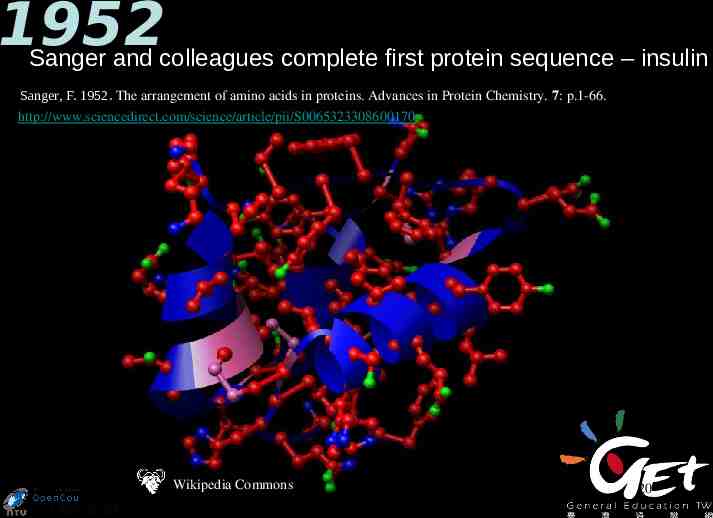
1952 Sanger and colleagues complete first protein sequence – insulin Sanger, F. 1952. The arrangement of amino acids in proteins. Advances in Protein Chemistry. 7: p.1-66. http://www.sciencedirect.com/science/article/pii/S0065323308600170 Wikipedia Commons 20

1953 Watson and Crick in 1953. Cold Spring Harbor Laboratory 21

1955 Smithies uses starch-gel electrophoresis to identify protein polymorphisms John C. Avise http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1215845/ Wikipedia Mapos 22

1963 Margoliash determines amino acid sequences for cytochrome c in several taxa and generates the first phylogenetic tree for a specific gene product. John C. Avise http://www.ncbi.nlm.nih.gov/pmc/articles/PMC221244/ 23

1965 Zuckerkandl and Pauling “molecular clock” hypothesis. http://en.wikipedia.org/wiki/History of molecular evolutio n 24
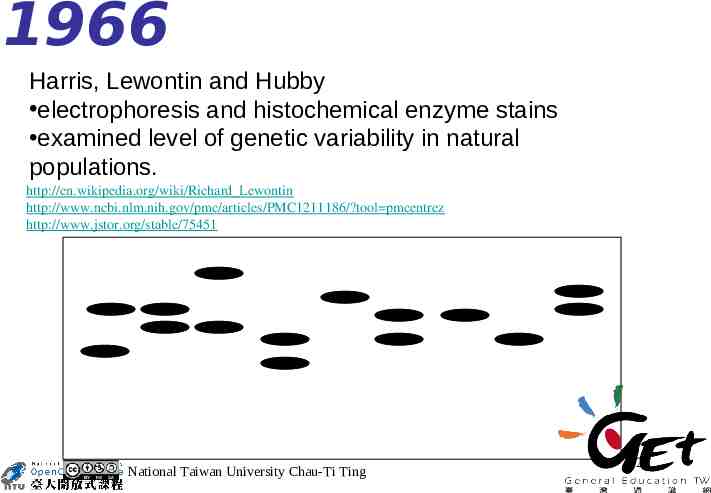
1966 Harris, Lewontin and Hubby electrophoresis and histochemical enzyme stains examined level of genetic variability in natural populations. http://en.wikipedia.org/wiki/Richard Lewontin http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1211186/?tool pmcentrez http://www.jstor.org/stable/75451 National Taiwan University Chau-Ti Ting 25
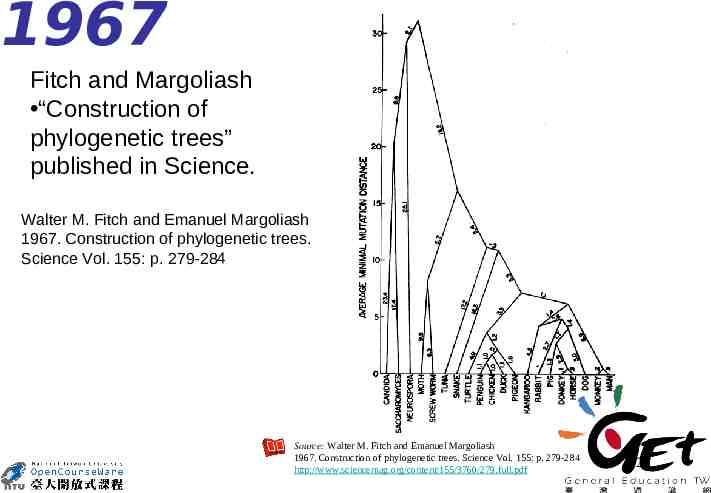
1967 Fitch and Margoliash “Construction of phylogenetic trees” published in Science. Walter M. Fitch and Emanuel Margoliash 1967. Construction of phylogenetic trees. Science Vol. 155: p. 279-284 Source: Walter M. Fitch and Emanuel Margoliash 1967. Construction of phylogenetic trees. Science Vol. 155: p. 279-284 http://www.sciencemag.org/content/155/3760/279.full.pdf 26

Source: Walter M. Fitch. 1988. Citation Classic. No. 27. http://garfield.library.upenn.edu/classics.html http://garfield.library.upenn.edu/classics1988/A1988N888200001.pdf 27

1968 Kimura proposes that the majority of molecular changes in evolution are due to random drift of neutral or nearly neutral mutations. http://en.wikipedia.org/wiki/Motoo Kimura#cite note-Kimura68-1 http://www.nature.com/nature/journal/v217/n5129/abs/217624a0.html Source: John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. 1969 King and Jukes “Non-Darwinian evolution” Also proposed neutral theory . http://en.wikipedia.org/wiki/Non-Darwinian Evolution http://www.sciencemag.org/content/164/3881/788.citation 28

Marshall Nirenberg, a young biochemist at the National Institute of Arthritic and Metabolic Diseases, discovered the first "triplet"—a sequence of three bases of DNA that codes for one of the twenty amino acids that serve as the building blocks of proteins. Subsequently, within five years, the entire genetic Genome News Network code was deciphered. Source: http://www.genomenewsnetwork.org/resources/timeline/1961 Nirenberg.php Cold Spring Harbor Laboratory Cold Spring Harbor Laboratory 29
1972 Nei’s genetic distance has greatly facilitated the study of evolutionary relationships among populations or closely http://en.wikipedia.org/wiki/Masatoshi Nei related species. http://www.jstor.org/stable/2459777 Source: M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 47. Discovery Publishing House. India. 30

1977 Molecular biologists by the 1970s had deciphered the genetic code and could spell out the sequence of amino acids in proteins. But inability to easily read off the precise nucleotide sequences of DNA forestalled further advances in molecular genetics and all prospects of genetic engineering. Walter Gilbert (with graduate student Allan M. Maxam) and Frederick Sanger, in 1977, working separately in the United States and England, developed new techniques for rapid DNA sequencing. Source: Genome News Network Cold Spring Harbor Laboratory 31

1981 Palmer and colleagues begin an important series papers utilizing cpDNA for phylogenetic reconstructions in plants Source: John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. 1984 Sibley and Ahlquist DNA-DNA hybridization primate evolution study. http://en.wikipedia.org/wiki/DNA-DNA hybridization http://www.springerlink.com/content/g3020651ml536640/ 32

Carolina Biological Supply Company Source: Carolina Biological Supply Company http://www.geneticorigins.org/ Science /AAAS Source: 2001. Science. Vol. 291. Issue 5507. http://www.sciencemag.org/content/291/5507.cover-expansion 33
Homework Assignment I 1) Write a paragraph about yourself (including name, background, and contact information) and your research interests. 2) What do you expect to learn from taking this course? Please send your homework as a pdf document online by Feb. 28th 34

Copyright Declaration Work P17. “The rates and patterns of changes in the genetic material and its encoded products during evolutionary time” P17. “The mechanisms responsible for such changes” P17. “The reconstruction of the evolutionary history of genes and organisms ” Licensing Author/Source M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 42. Discovery Publishing House. India. http://books.google.com.tw/books?id OoG4 y2Q9AEC&printsec frontcover&hl zh-TW#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 42. Discovery Publishing House. India. http://books.google.com.tw/books?id OoG4 y2Q9AEC&printsec frontcover&hl zh-TW#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 42. Discovery Publishing House. India. http://books.google.com.tw/books?id OoG4 y2Q9AEC&printsec frontcover&hl zh-TW#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Science /AAAS 2001. Science. Vol. 291. Issue 5507. http://www.sciencemag.org/content/291/5507.cover-expansion It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Science /AAAS Copyright Statement Carolina Biological Supply Company http://www.geneticorigins.org/ 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Carolina Biological Supply Company Terms of Use Page P17 P17 P17 P18, 33 P18, 35 33
Work P19. “Nuttall conducted precipitin tests of serum proteins to infer the phylogenetic relationship among various groups of animals.” P22. “Smithies uses starch-gel electrophoresis to identify protein polymorphisms.” Licensing Author/Source M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 43. Discovery Publishing House. India. http://books.google.com.tw/books?id OoG4 y2Q9AEC&printsec frontcover&hl z h-TW#v onepage&q&f false It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 P19 Wikipedia Commons http://commons.wikimedia.org/wiki/File:Insulin.jpg 2012/02/21 visited P20 Watson and Crick in 1953.Cold Spring Harbor Laboratory http://www.dnalc.org/view/16430-Gallery-19-DNA-model-1953.html 2012/02/21 visited P21 John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. http://books.google.com.tw/books?id aWSbZf-dRbEC&printsec frontcover&hl zh -TW#v onepage&q&f false It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Wikipedia Mapos http://en.wikipedia.org/wiki/File:Oliver-smithies.jpg 2012/02/22 visited P23. “Margoliash determines amino acid sequences for cytochrome c in several taxa and generates the first phylogenetic tree for a specific gene product.” Page John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. http://books.google.com.tw/books?id aWSbZf-dRbEC&printsec frontcover&hl zh -TW#v onepage&q&f false It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Open Clip Art Library Anonymous http://openclipart.org/detail/108235/view-tree-by-anonymous 2012/02/21 visited P22 P22 P23 P23, 30 36

Work Licensing Author/Source Page Wikipedia brian0918 http://en.wikipedia.org/wiki/File:ADN animation.gif 2012/02/21 visited P24 Open Clip Art Library fzap http://openclipart.org/detail/16605/clock-sportstudio-design-by-fzap 2012/02/21 visited P24 National Taiwan University Chau-Ti Ting P25 Walter M. Fitch and Emanuel Margoliash. 1967. Construction of phylogenetic trees. Science Vol. 155: p.282 http://www.sciencemag.org/content/155/3760/279.extract?sid fe0a324a-4d85-453e -b77a-8a69782df245 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Science /AAAS Copyright Statement Walter M. Fitch 1988. Citation Classic. No. 27. http://garfield.library.upenn.edu/classics.html http://garfield.library.upenn.edu/classics1988/A1988N888200001.pdf 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 The "Code of Best Practices in Fair Use for OpenCourseWare 2009 ( http://www.centerforsocialmedia.org/sites/default/files/10-305-OCW-Oct29.pdf)" by A Committee of Practitioners of OpenCourseWare in the U.S. The contents are based on Section 107 of the 1976 U.S. Copyright Act P26 P27 37
Work P28. “Kimura proposes that the majority of molecular changes in evolution are due to random drift of neutral or nearly neutral mutations.” P29. “Marshall Nirenberg, a young biochemist at the National Institute of Arthritic and Metabolic Diseases Subsequently, within five years, the entire genetic code was deciphered” P30. “Nei’s genetic distance has greatly facilitated the study of evolutionary relationships among populations or closely related species.” P31. “Molecular biologists by the 1970s had deciphered developed new techniques for rapid DNA sequencing.” Licensing Author/Source John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. http://books.google.com.tw/books?id aWSbZf-dRbEC&printsec frontcover&hl zh-T W#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Page P28 Genome News Network http://www.genomenewsnetwork.org/resources/timeline/1961 Nirenberg.php 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 P29 Cold Spring Harbor Laboratory http://www.dnalc.org/view/16497-Gallery-22-Marshall-Nirenberg-1960.html 2012/02/21 visited P29 Cold Spring Harbor Laboratory http://www.dnalc.org/view/16498-Gallery-22-Marshall-Nirenberg-1999.html 2012/02/21 visited P29 M. Prakash 2007. Encyclopaedia of Gene Evolution Vol. 2, Molecular Genetics, p. 47. Discovery Publishing House. India. http://books.google.com.tw/books?id OoG4 y2Q9AEC&printsec frontcover&hl zhTW#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 Genome News Network http://www.genomenewsnetwork.org/resources/timeline/1977 Gilbert.php 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 P30 P31 38
Work P32. “Palmer and colleagues begin an important series papers utilizing cpDNA for phylogenetic reconstructions in plants” Licensing Author/Source Page Cold Spring Harbor Laboratory http://www.dnalc.org/view/16520-Gallery-23-Fred-Sanger-late-1950-s.html 2012/02/21 visited P31 John C. Avise 1994. Molecular Markers, Natural History and Evolution, p. 42. Chapman & Hall. New York, USA. http://books.google.com.tw/books?id aWSbZf-dRbEC&printsec frontcover&hl zhTW#v onepage&q&f false 2012/02/21 visited It is used subject to the fair use doctrine of: Taiwan Copyright Act Articles 52 & 65 P32 39






