Structure-based Drug Design
19 Slides992.50 KB

Structure-based Drug Design
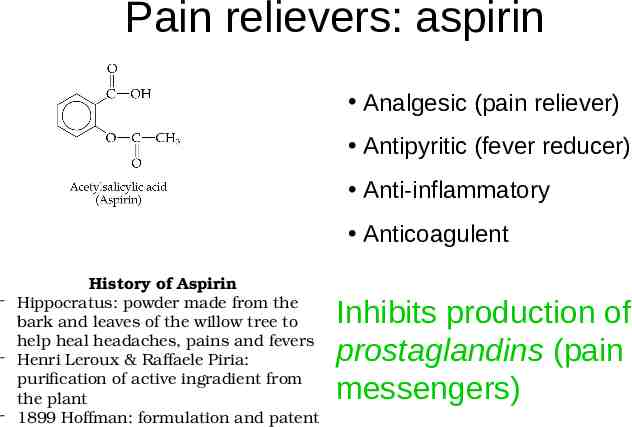
Pain relievers: aspirin Analgesic (pain reliever) Antipyritic (fever reducer) Anti-inflammatory Anticoagulent History of Aspirin - Hippocratus: powder made from the bark and leaves of the willow tree to help heal headaches, pains and fevers - Henri Leroux & Raffaele Piria: purification of active ingradient from the plant - 1899 Hoffman: formulation and patent Inhibits production of prostaglandins (pain messengers)
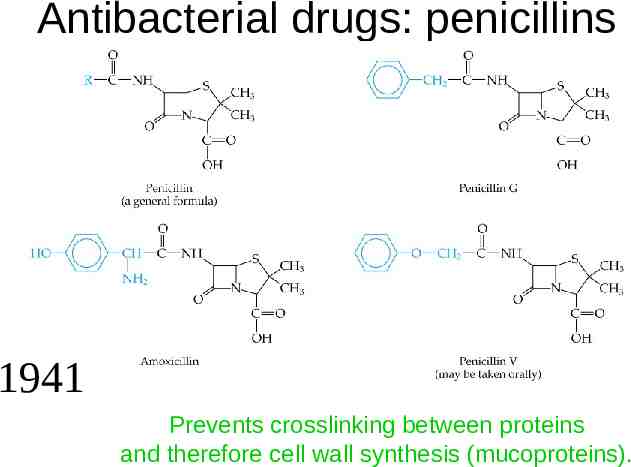
Antibacterial drugs: penicillins 1941 Prevents crosslinking between proteins and therefore cell wall synthesis (mucoproteins).
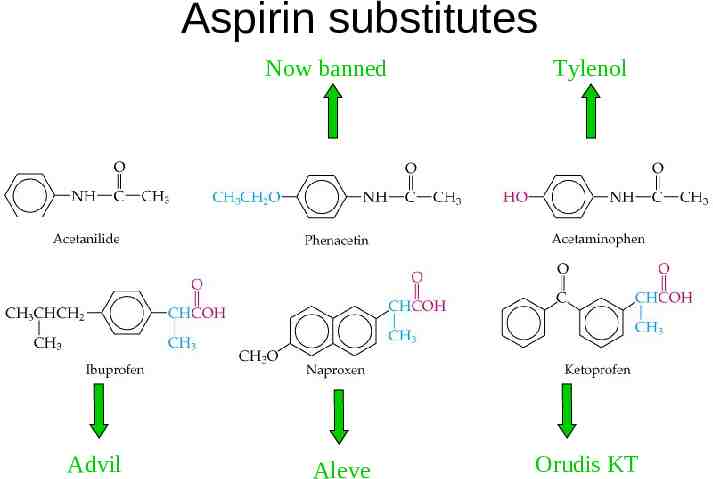
Aspirin substitutes Now banned Advil Aleve Tylenol Orudis KT
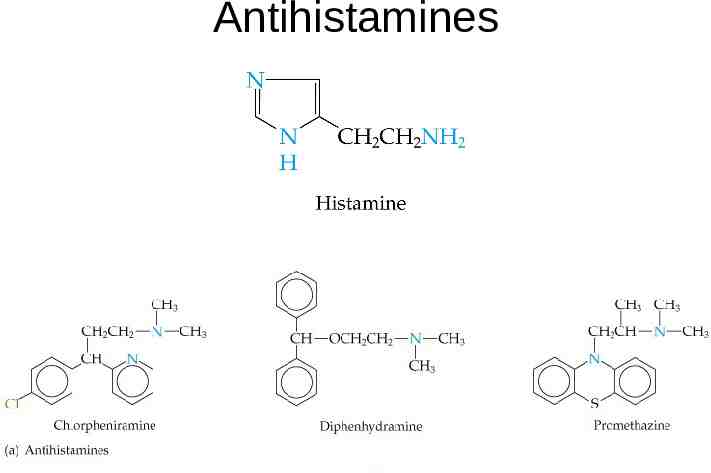
Antihistamines
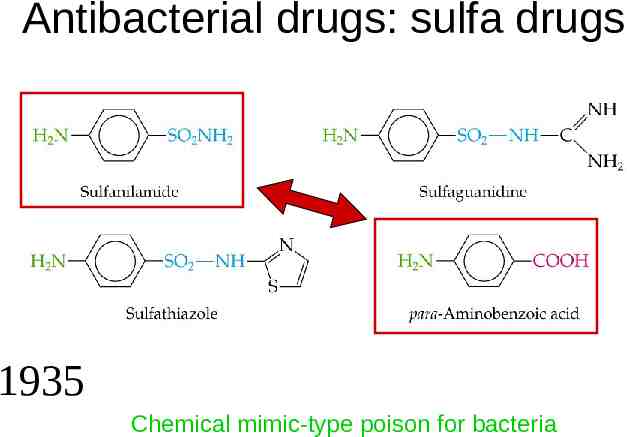
Antibacterial drugs: sulfa drugs 1935 Chemical mimic-type poison for bacteria
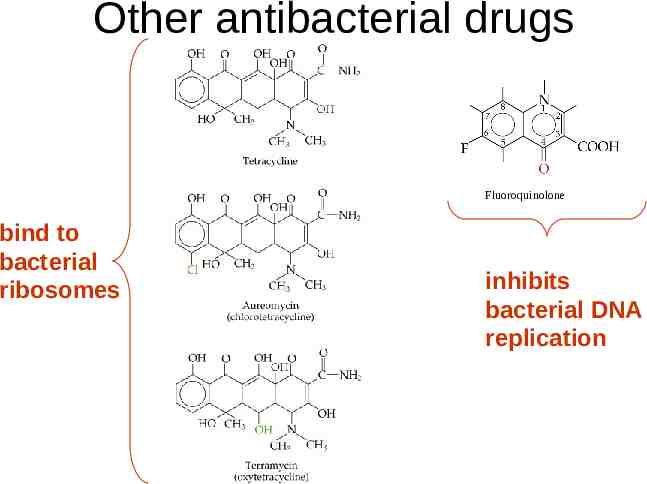
Other antibacterial drugs Fluoroquinolone bind to bacterial ribosomes inhibits bacterial DNA replication
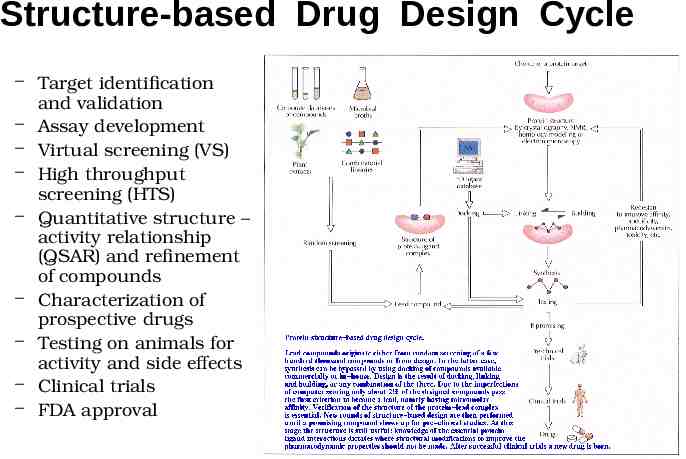
Structure-based Drug Design Cycle – Target identification and validation – Assay development – Virtual screening (VS) – High throughput screening (HTS) – Quantitative structure – activity relationship (QSAR) and refinement of compounds – Characterization of prospective drugs – Testing on animals for activity and side effects – Clinical trials – FDA approval
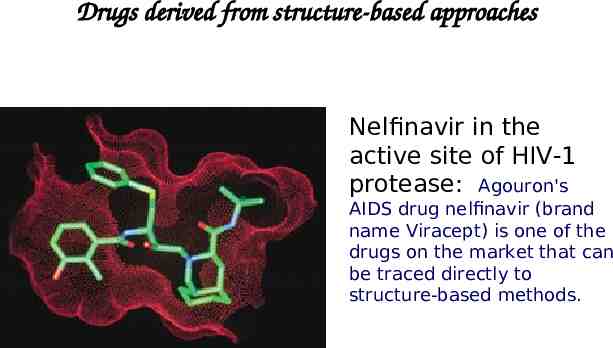
Drugs derived from structure-based approaches Nelfinavir in the active site of HIV-1 protease: Agouron's AIDS drug nelfinavir (brand name Viracept) is one of the drugs on the market that can be traced directly to structure-based methods.
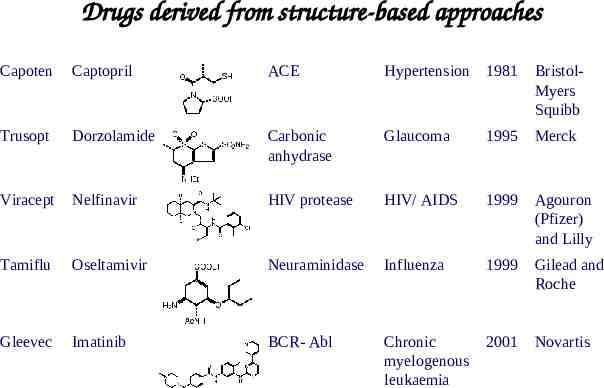
Drugs derived from structure-based approaches Capoten Captopril ACE Hypertension 1981 BristolMyers Squibb Trusopt Dorzolamide Carbonic anhydrase Glaucoma 1995 Merck Viracept Nelfinavir HIV protease HIV/ AIDS 1999 Agouron (Pfizer) and Lilly Tamiflu Oseltamivir Neuraminidase Influenza 1999 Gilead and Roche Gleevec Imatinib BCR- Abl Chronic myelogenous leukaemia 2001 Novartis

Determination of Target Structure Crystal structure of Rhodopsin: A G proteincoupled receptor. Palczewski et al. Science (2000) 289, 739- 45.
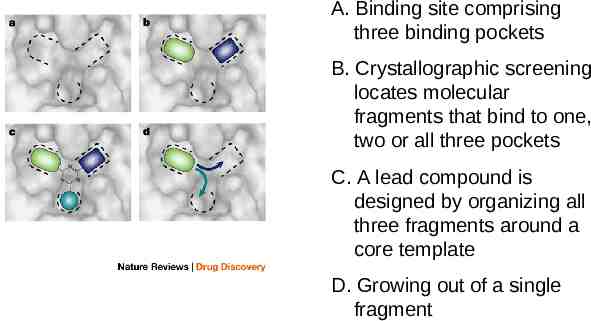
A. Binding site comprising three binding pockets B. Crystallographic screening locates molecular fragments that bind to one, two or all three pockets C. A lead compound is designed by organizing all three fragments around a core template D. Growing out of a single fragment
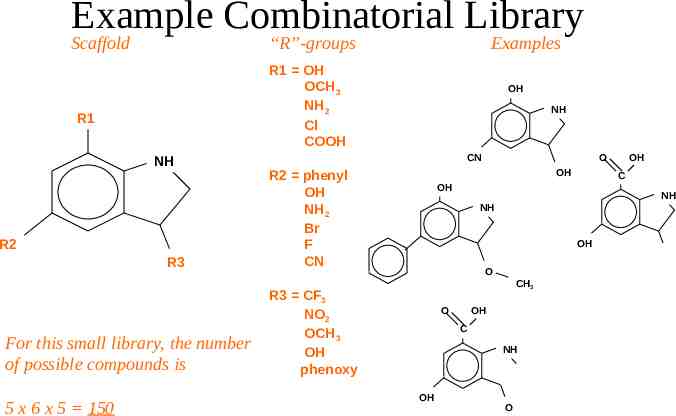
Example Combinatorial Library Scaffold “R”-groups R1 OH OCH3 NH2 Cl COOH R1 NH R2 R3 For this small library, the number of possible compounds is 5 x 6 x 5 150 Examples OH NH CN R2 phenyl OH NH2 Br F CN O OH OH C OH NH NH OH O CH3 R3 CF3 NO2 OCH3 OH phenoxy O OH C NH OH O CF3
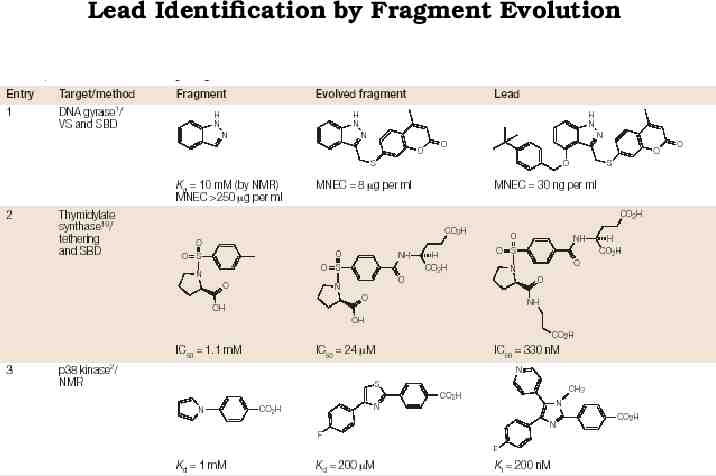
Lead Identification by Fragment Evolution
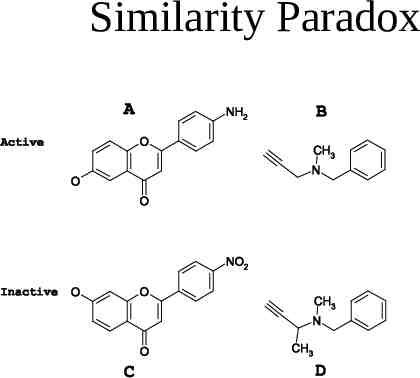
Similarity Paradox

Descriptors of Molecular Structure & Properties 1D-descriptors encode chemical composition & physicochemical properties – MW, CmOnHk ,hydrophobicity 2D-descriptors encode chemical topology – Connectivity indices, degree of branching, degree of flexibility, # of aromatic bonds 3D-descriptors encode 3D shape, volume, functionality, surface area – Pharmacophore – the spatial arrangement of chemical groups that determines its activity

Lipinski Rule of Five (1997) Poor absorption and permeation are more likely to occur when there are more than 5 hydrogenbond donors, more than 10 hydrogen-bond acceptors, the molecular mass is greater than 500, or the log P value is greater than 5. Further research studied a broader range of physicochemical and structural properties Related problems: – – – – Compound toxicity Compound mutagenicity Blood-brain barrier penetration Central nervous system activity
In Silico ADME Models Computational methods can predict compound properties important to ADME, e.g. – – – – LogP, a liphophilicity measure Solubility Permeability Cytochrome p450 metabolism Means estimates can be made for millions of compouds, helping reduce “attrition” – the failure rate of compounds in late stage
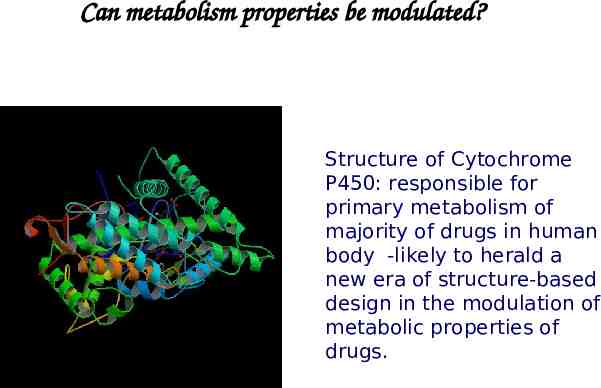
Can metabolism properties be modulated? Structure of Cytochrome P450: responsible for primary metabolism of majority of drugs in human body -likely to herald a new era of structure-based design in the modulation of metabolic properties of drugs.






