Molecule as Computation Ehud Shapiro Weizmann Institute of Science
69 Slides8.37 MB

Molecule as Computation Ehud Shapiro Weizmann Institute of Science Joint work with Aviv Regev and Bill Silverman In collaboration with Corrado Priami, Naama Barkai and Luca Cardelli

The talk has three parts: 1. Briefly introduce molecular biology 2. Computer-based consolidation of molecular biology 3. Our work on helping this happen

Part I Brief Introduction to Molecular Biology
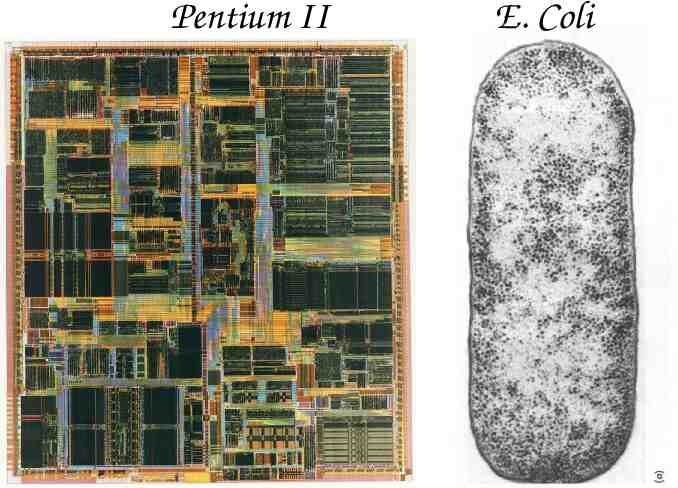
Pentium II E. Coli
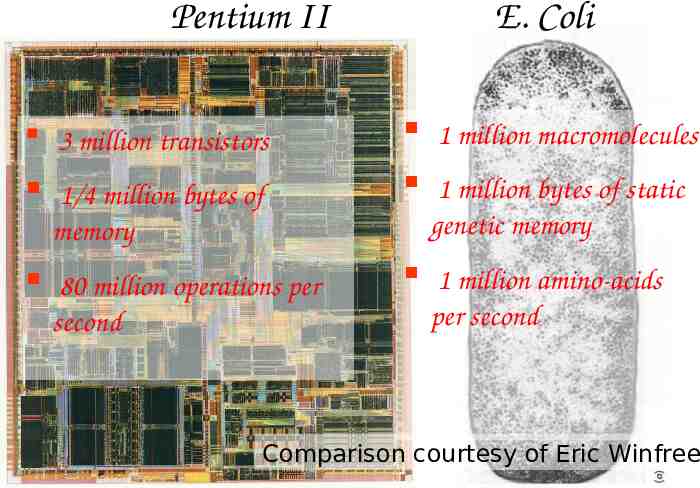
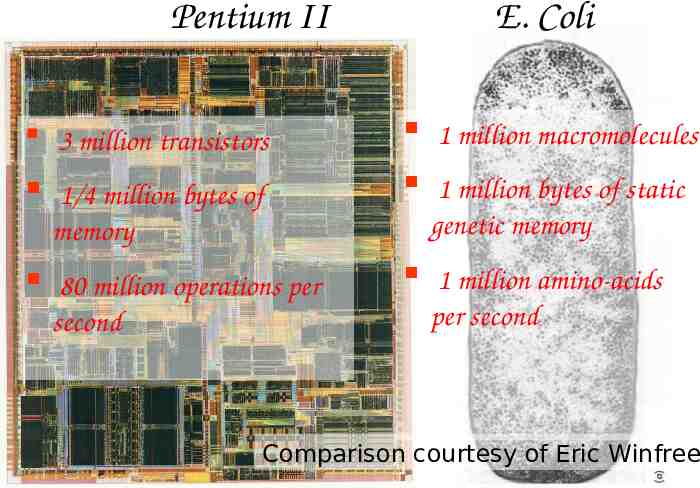
Pentium II 3 million transistors 1/4 million bytes of memory 80 million operations per second E. Coli 1 million macromolecules 1 million bytes of static genetic memory 1 million amino-acids per second Comparison courtesy of Eric Winfree
Pentium II E. Coli
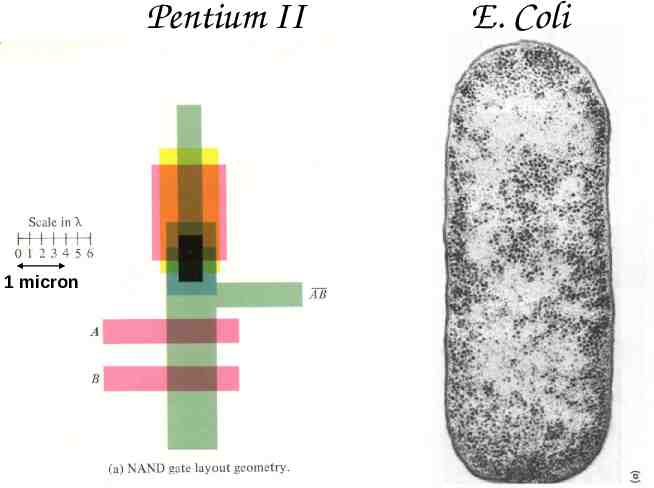
Pentium II 1 micron E. Coli
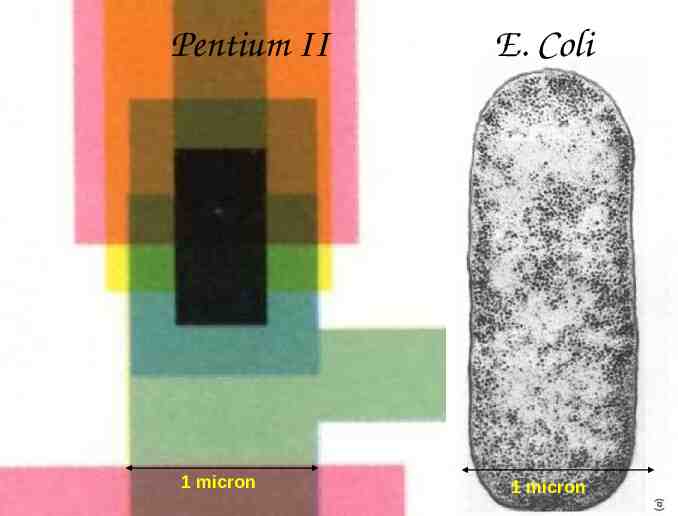
Pentium II 1 micron E. Coli 1 micron
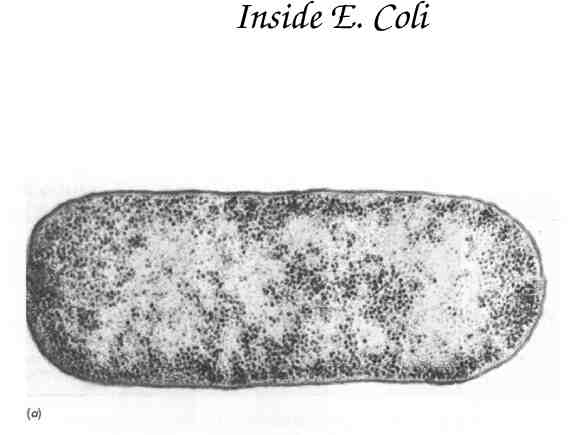
Inside E. Coli
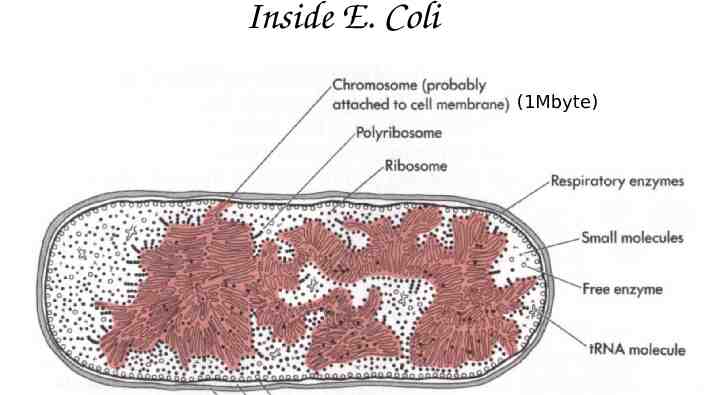
Inside E. Coli (1Mbyte)
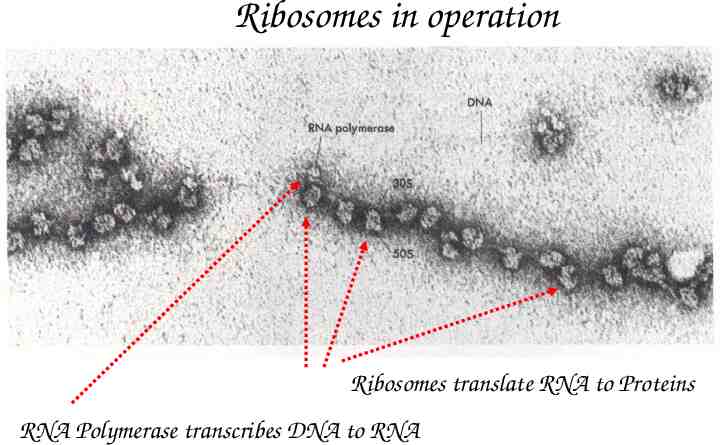
Ribosomes in operation Ribosomes translate RNA to Proteins RNA Polymerase transcribes DNA to RNA
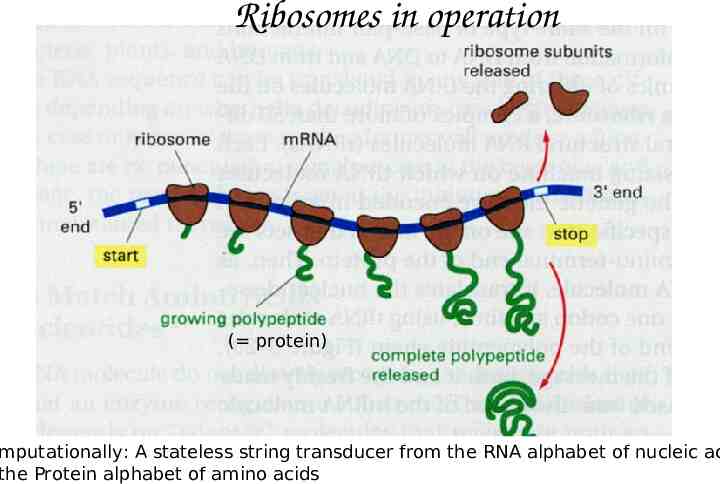
Ribosomes in operation ( protein) mputationally: A stateless string transducer from the RNA alphabet of nucleic ac the Protein alphabet of amino acids
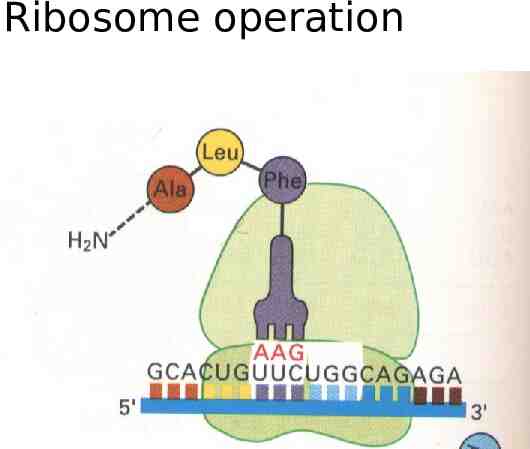
Ribosome operation
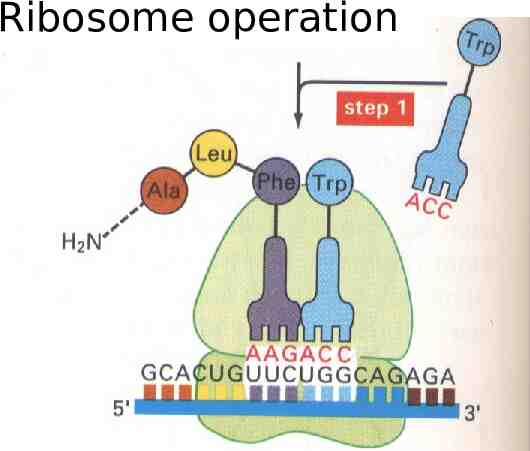
Ribosome operation
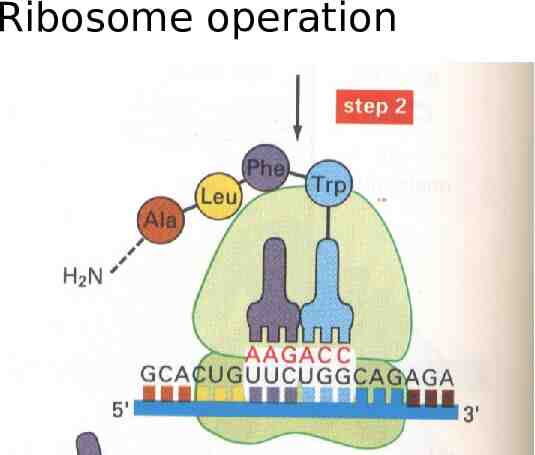
Ribosome operation
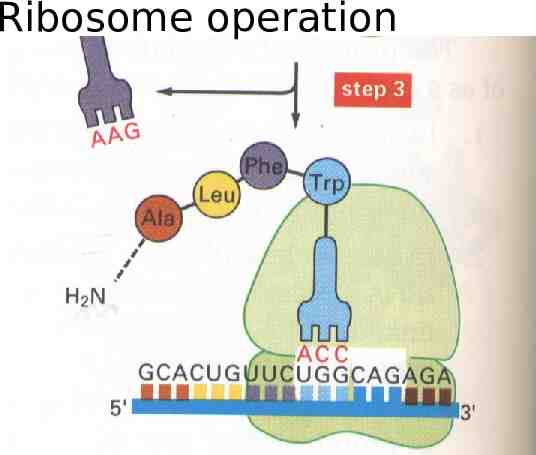
Ribosome operation
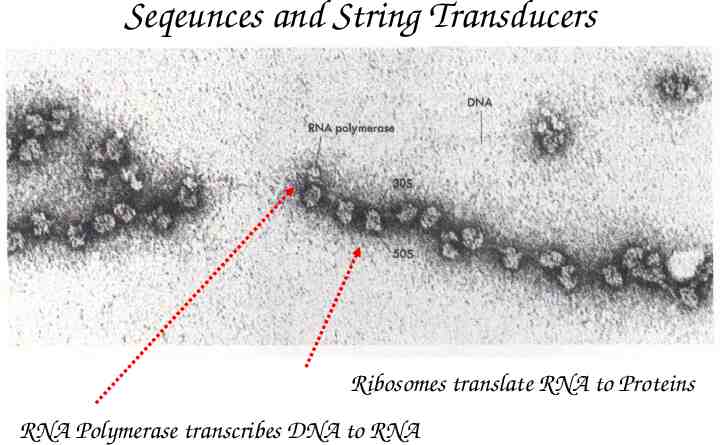
Seqeunces and String Transducers Ribosomes translate RNA to Proteins RNA Polymerase transcribes DNA to RNA

Molecular Biology in One Slide Sequence: Sequence of DNA and Proteins

Molecule as Computation Ehud Shapiro Weizmann Institute of Science Joint work with Aviv Regev and Bill Silverman In collaboration with Corrado Priami, Naama Barkai and Luca Cardelli
The talk has three parts: 1. Briefly introduce molecular biology 2. Computer-based consolidation of molecular biology 3. Our work on helping this happen

Part I Brief Introduction to Molecular Biology

Pentium II E. Coli

Pentium II 3 million transistors 1/4 million bytes of memory 80 million operations per second E. Coli 1 million macromolecules 1 million bytes of static genetic memory 1 million amino-acids per second Comparison courtesy of Eric Winfree
What about “The Rest” of biology: the function, activity and interaction of molecular systems in cells? ?
Part III An Abstraction for Molecular Systems

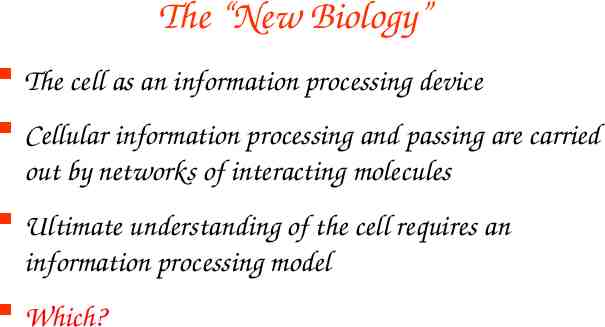
The “New Biology” The cell as an information processing device Cellular information processing and passing are carried out by networks of interacting molecules Ultimate understanding of the cell requires an information processing model Which?

“We have no real ‘algebra’ for describing regulatory circuits across different systems.” - T. F. Smith (TIG 14:291-293, 1998) “The data are accumulating and the computers are humming, what we are lacking are the words, the grammar and the syntax of a new language ” - D. Bray (TIBS 22:325-326, 1997)
Our Proposal: Molecule as Computational Process A system of interacting molecular entities is described and modelled by a system of interacting computational entities. “Cellular Abstractions: Cells as Computation”, to appear in Nature, September 26th, 2002

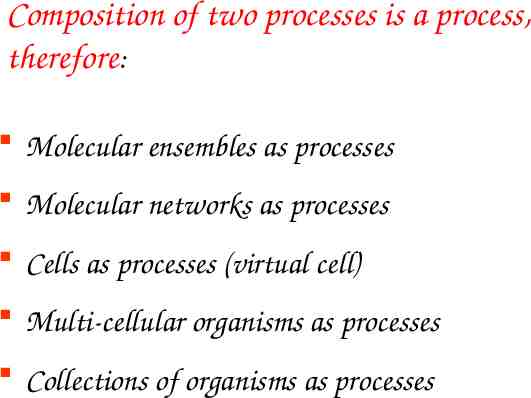
Composition of two processes is a process, therefore: Molecular ensembles as processes Molecular networks as processes Cells as processes (virtual cell) Multi-cellular organisms as processes Collections of organisms as processes

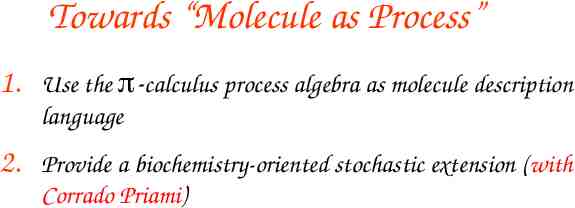
Towards “Molecule as Process” 1. Use the -calculus process algebra as molecule description language
The -calculus (Milner, Walker and Parrow 1989) A program specifies a network of interacting processes Communication occurs on complementary channels, identified by names Message content: Channel name Processes are defined by their potential communication activities

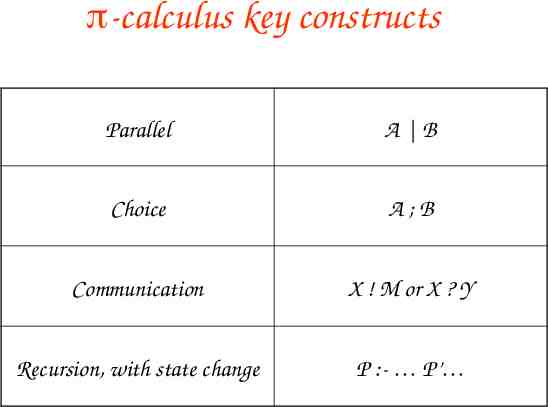
-calculus key constructs Parallel A B Choice A;B Communication X ! M or X ? Y Recursion, with state change P :- P’

Molecules as Processes Molecule Process Interaction capability Channel Interaction Communication Modification State change
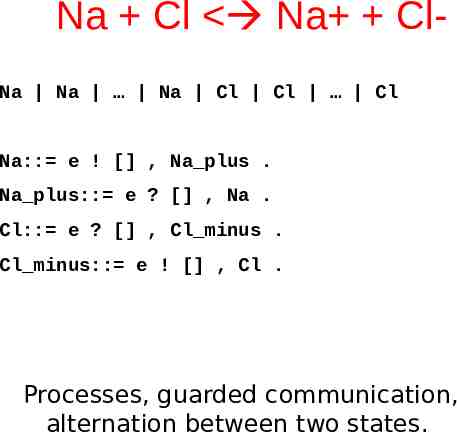
Na Cl Na ClNa Na Na Cl Cl Cl Na:: e ! [] , Na plus . Na plus:: e ? [] , Na . Cl:: e ? [] , Cl minus . Cl minus:: e ! [] , Cl . Processes, guarded communication, alternation between two states.

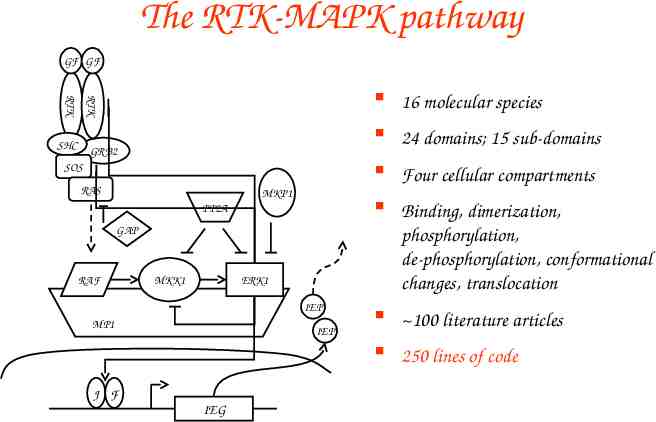
The RTK-MAPK pathway GF GF RTK RTK SHC GRB2 SOS RAS MKP1 PP2A GAP RAF MKK1 ERK1 IEP MP1 IEP J F IEG 16 molecular species 24 domains; 15 sub-domains Four cellular compartments Binding, dimerization, phosphorylation, de-phosphorylation, conformational changes, translocation 100 literature articles 250 lines of code
Molecular systems with -calculus Can express, qualitatively, the behavior of many complex molecular systems Cannot express quantitative aspects
Towards “Molecule as Process” 1. Use the -calculus process algebra as molecule description language 2. Provide a biochemistry-oriented stochastic extension (with Corrado Priami)

Stochastic -Calculus (Priami, 1995, Regev, Priami, Shapiro, Silverman 2000) Every channel x attached with a base rate r A global (external) clock is maintained The clock is advanced and a communication is selected according to a race condition Rate calculation and race condition adapted for chemical reactions: Rate(A B C) BaseRate *[A]*[B] [A] number of A’s willing to communicate with B’s. [B] number of B’s willing to communicate with A’s.
BioSPI implementation: -calculus Gillespie’s algorithm Gillespie (1977): Accurate stochastic simulation of chemical reactions The BioSPI system: Compiles (full) calculus Runtime incorporates Gillespie’s algorithm

Na Cl Na Cl100 90 80 global(e1(100),e2(10)). 70 60 50 40 30 Na:: e1 ! [] , Na plus . 20 Na plus:: e2 ? [] , Na . 0 Cl:: e1 ? [] , Cl minus . 10 0 0.5 1 1.5 2 2.5 3 3.5 100 4 x 10 -3 90 80 70 Cl minus:: e2 ! [] , Cl . 60 50 40 30 20 10 0 0 0.005 0.01 0.015 0.02 0.025 0.03

Programming Experience with Stochastic Pi Calculus Taught semesterial M.Sc. Course (available online) with lots of examples, exercises and final projects Textbook examples from chemistry, organic chemistry, enzymatic reactions, metabolic pathways, signal-transduction pathways
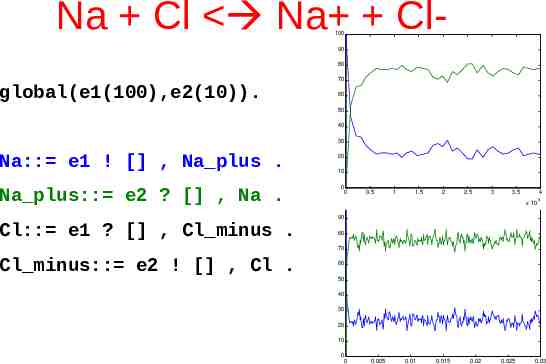
Circadian Clocks J. Dunlap, Science (1998) 280 1548-9

The circadian clock machinery (Barkai and Leibler, Nature 2000) A R degradation A R translation UTRA transcription UTRR A RNA PA PR transcription A GENE Differential rates: Very fast, fast and slow degradation translation R RNA R GENE
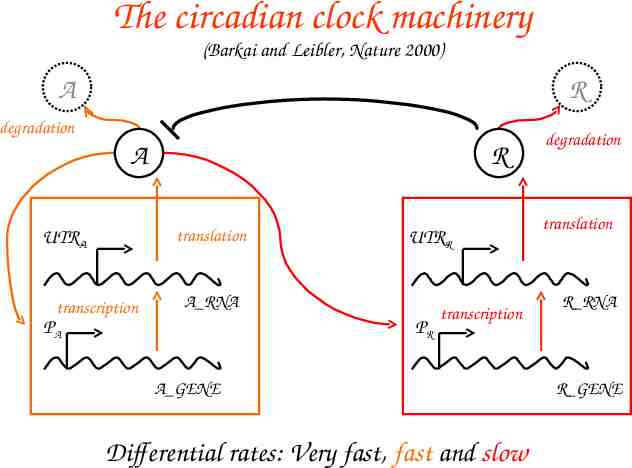
The machinery in -calculus: “A” molecules A GENE:: PROMOTED A BASAL A PROMOTED A:: pA ? {e}.ACTIVATED TRANSCRIPTION A(e) BASAL A:: bA ? [].( A GENE A RNA) ACTIVATED TRANSCRIPTION A:: 1 . (ACTIVATED TRANSCRIPTION A A RNA) e ? [] . A GENE RNA A:: TRANSLATION A DEGRADATION mA TRANSLATION A:: utrA ? [] . (A RNA A PROTEIN) DEGRADATION mA:: degmA ? [] . 0 A Gene A RNA A PROTEIN:: (new e1,e2,e3) PROMOTION A-R BINDING R DEGRADATION A PROMOTION A-R :: pA!{e2}.e2![]. A PROTEIN pR!{e3}.e3![]. A PRTOEIN BINDING R :: rbs ! {e1} . BOUND A PRTOEIN BOUND A PROTEIN:: e1 ? [].A PROTEIN degpA ? [].e1 ![].0 DEGRADATION A:: degpA ? [].0 A protein
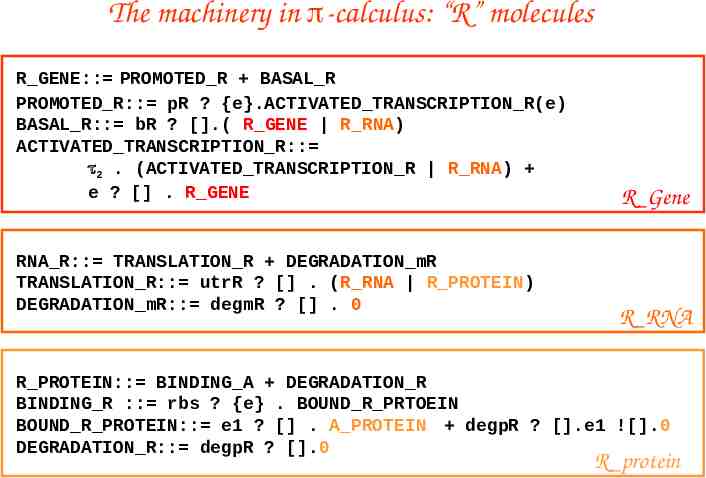
The machinery in -calculus: “R” molecules R GENE:: PROMOTED R BASAL R PROMOTED R:: pR ? {e}.ACTIVATED TRANSCRIPTION R(e) BASAL R:: bR ? [].( R GENE R RNA) ACTIVATED TRANSCRIPTION R:: 2 . (ACTIVATED TRANSCRIPTION R R RNA) e ? [] . R GENE RNA R:: TRANSLATION R DEGRADATION mR TRANSLATION R:: utrR ? [] . (R RNA R PROTEIN) DEGRADATION mR:: degmR ? [] . 0 R Gene R RNA R PROTEIN:: BINDING A DEGRADATION R BINDING R :: rbs ? {e} . BOUND R PRTOEIN BOUND R PROTEIN:: e1 ? [] . A PROTEIN degpR ? [].e1 ![].0 DEGRADATION R:: degpR ? [].0 R protein
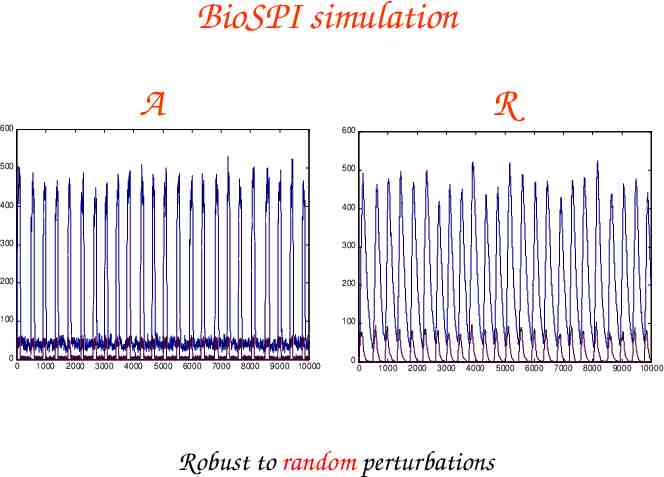
BioSPI simulation A 600 600 500 500 400 400 300 300 200 200 100 100 0 0 1000 2000 3000 4000 5000 R 6000 7000 8000 9000 10000 0 0 1000 2000 3000 4000 Robust to random perturbations 5000 6000 7000 8000 9000 10000
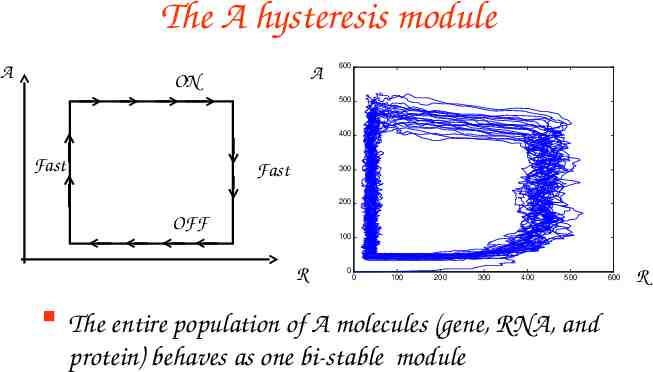
The A hysteresis module A A ON 600 500 400 Fast Fast 300 200 OFF 100 R 0 0 100 200 300 400 500 The entire population of A molecules (gene, RNA, and protein) behaves as one bi-stable module 600 R
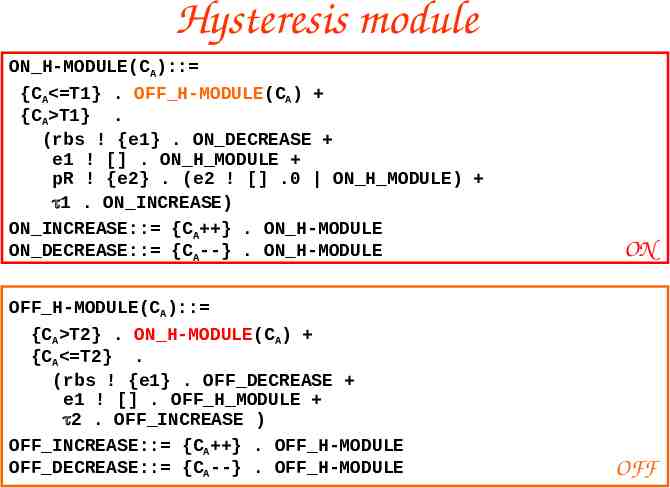
Hysteresis module ON H-MODULE(CA):: {CA T1} . OFF H-MODULE(CA) {CA T1} . (rbs ! {e1} . ON DECREASE e1 ! [] . ON H MODULE pR ! {e2} . (e2 ! [] .0 ON H MODULE) 1 . ON INCREASE) ON INCREASE:: {CA } . ON H-MODULE ON DECREASE:: {CA--} . ON H-MODULE OFF H-MODULE(CA):: {CA T2} . ON H-MODULE(CA) {CA T2} . (rbs ! {e1} . OFF DECREASE e1 ! [] . OFF H MODULE 2 . OFF INCREASE ) OFF INCREASE:: {CA } . OFF H-MODULE OFF DECREASE:: {CA--} . OFF H-MODULE ON OFF
Modular cell biology Build two representations in the -calculus Implementation (how?): molecular level Specification (what?): functional module level
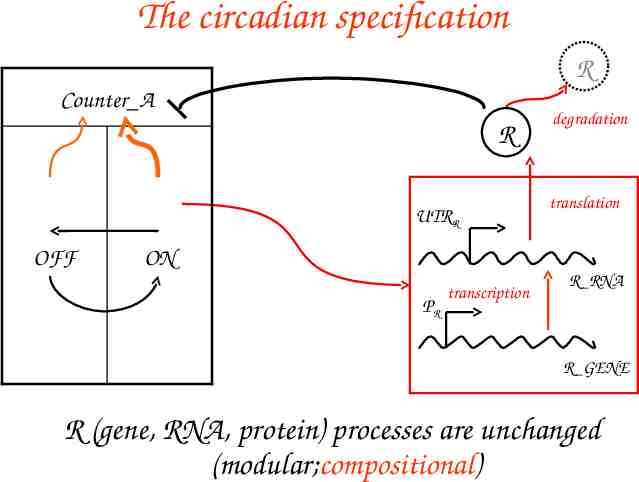
The circadian specification R Counter A R UTRR OFF degradation translation ON PR transcription R RNA R GENE R (gene, RNA, protein) processes are unchanged (modular;compositional)
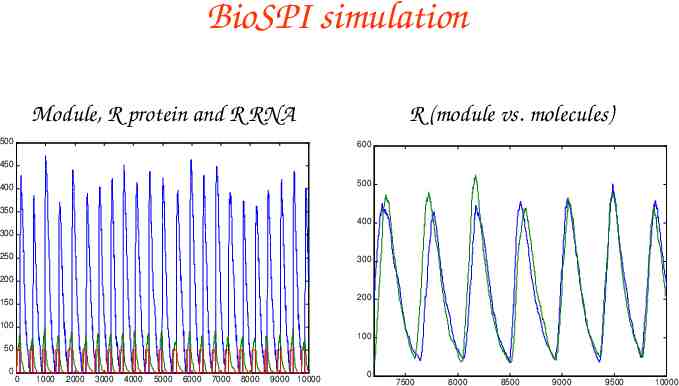
BioSPI simulation Module, R protein and R RNA 500 R (module vs. molecules) 600 450 500 400 350 400 300 250 300 200 200 150 100 100 50 0 0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0 7500 8000 8500 9000 9500 10000
Modular cell biology Build two representations in the -calculus Implementation (how?): molecular level Specification (what?): functional module level Ascribing a function to a biomolecular system equivalence between specification and implementation
Limitation of stochastic - calculus: Lack of location information Membranes: Cells and cellular compartments, “inside” and “outside” Molecular proximity: The identity of complexes and single molecules Limited solution: programming tricks
Towards “Molecule as Process” 1. Use the -calculus process algebra as molecule description language 2. Provide a biochemistry-oriented stochastic extension (with Corrado Priami) 3. Provide an Ambient Calculus extension (with Luca Cardelli)

Mobile compartments Compartment Compartment mobility Process mobility Cells Cell movement Organelles and vesicles Merging, budding, bursting Trans-membranal molecules (receptors, channels, transporters); Multi-molecular complexes Form and break Molecule entry and exit Bind and unbind to molecular scaffolds
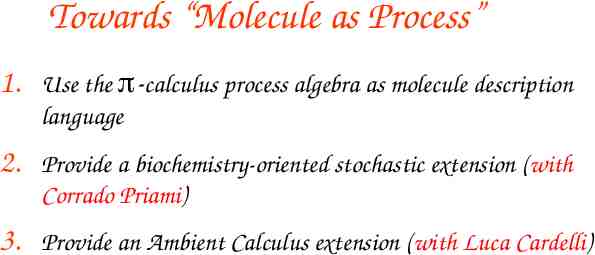
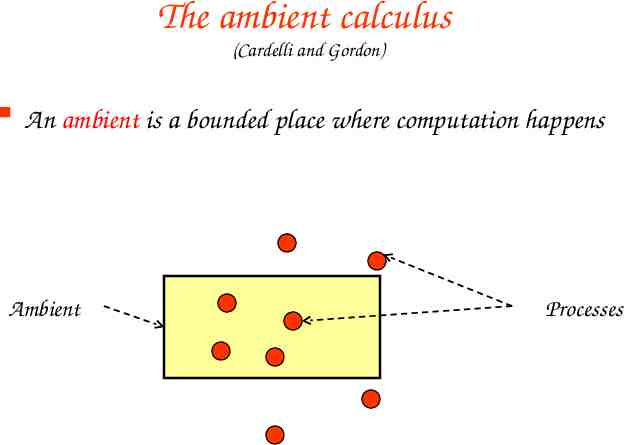
The ambient calculus (Cardelli and Gordon) An ambient is a bounded place where computation happens Ambient Processes
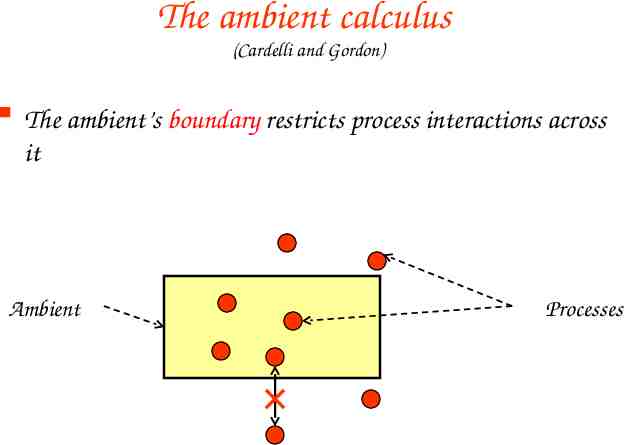
The ambient calculus (Cardelli and Gordon) The ambient’s boundary restricts process interactions across it Ambient Processes
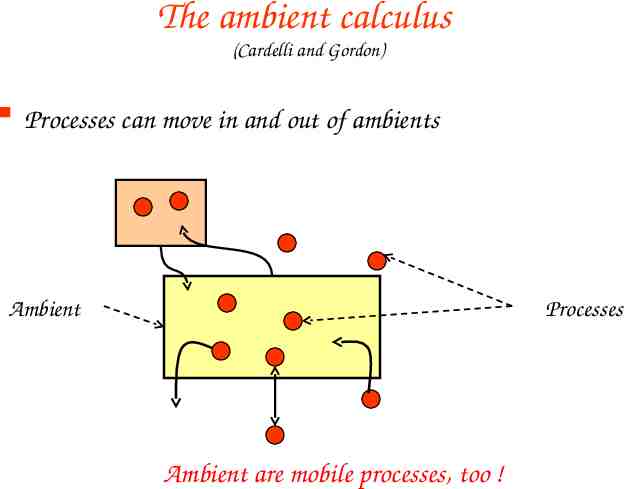
The ambient calculus (Cardelli and Gordon) Processes can move in and out of ambients Ambient Processes Ambient are mobile processes, too !
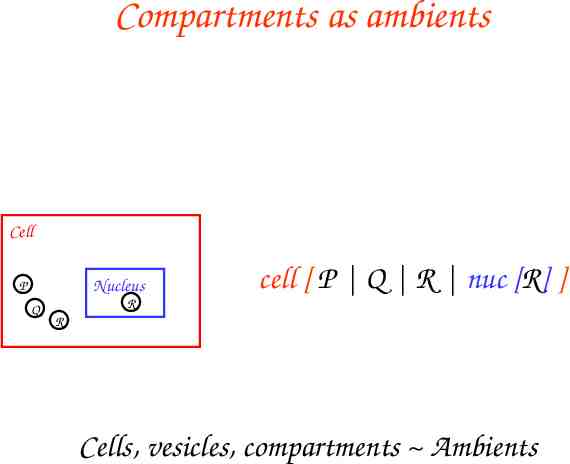
Compartments as ambients Cell Nucleus P Q R cell [ P Q R nuc [R] ] R Cells, vesicles, compartments Ambients
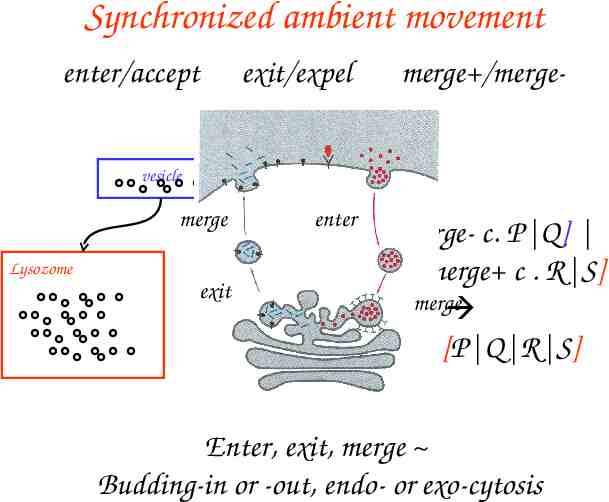
Synchronized ambient movement enter/accept exit/expel merge /merge- vesicle merge Lysozome exit enter vesicle[merge- c. P Q] lysozome [merge c . R S] merge lysozome [P Q R S] Enter, exit, merge Budding-in or -out, endo- or exo-cytosis
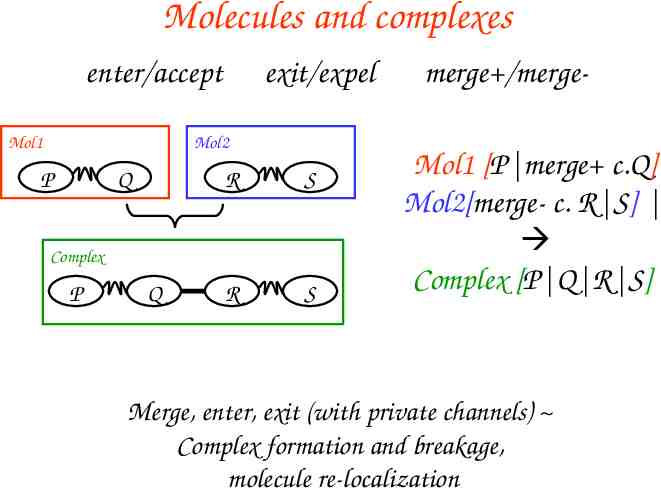
Molecules and complexes enter/accept Mol1 exit/expel Mol2 P Q R S R S Complex P Q merge /mergeMol1 [P merge c.Q] Mol2[merge- c. R S] Complex [P Q R S] Merge, enter, exit (with private channels) Complex formation and breakage, molecule re-localization
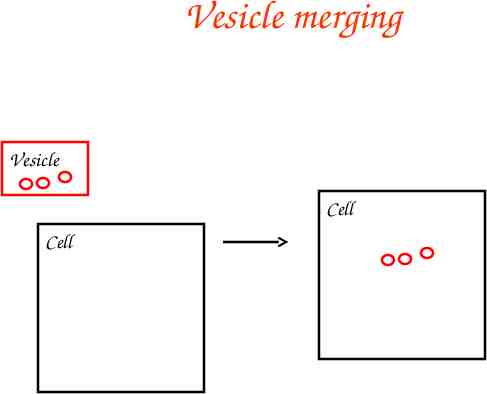
Vesicle merging Vesicle Cell Cell
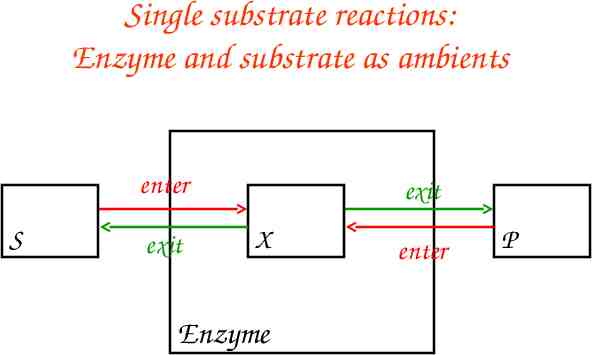
Single substrate reactions: Enzyme and substrate as ambients enter S exit exit X Enzyme enter P
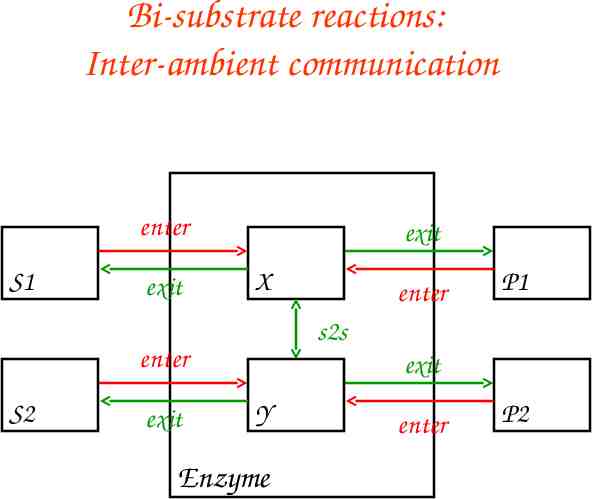
Bi-substrate reactions: Inter-ambient communication enter S1 exit exit X exit P1 s2s enter S2 enter exit Y Enzyme enter P2
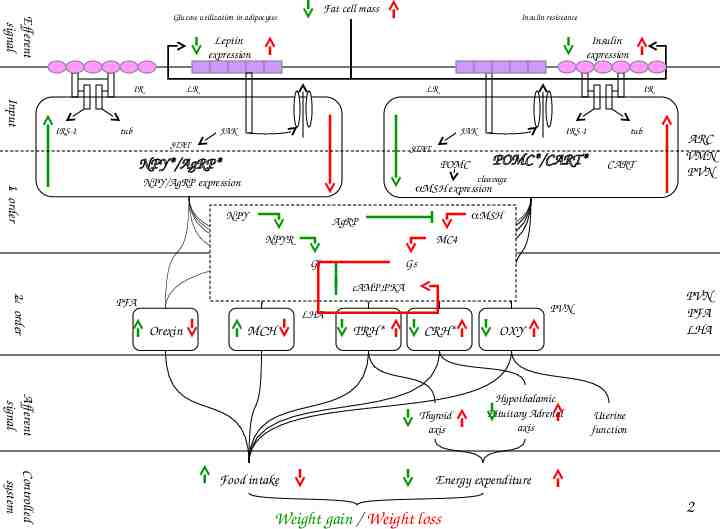
Example: Multi-cellular system (hypothalamic body weight control system)
Fat cell mass Efferent signal Glucose utilization in adipocytes Insulin resistance Leptin expression IR Insulin expression LR LR IR Input IRS-1 tub JAK JAK STAT IRS-1 STAT NPY*/AgRP* POMC*/CART* POMC tub CART cleavage 1st order NPY/AgRP expression MSH expression NPY MSH AgRP NPYR MC4 Gi Gs 2nd order cAMP,PKA PFA MCH TRH* CRH* Afferent signal Thyroid axis Controlled system Food intake PVN PFA LHA PVN LHA Orexin ARC VMN PVN OXY Hypothalamic Pituitary Adrenal axis Uterine function Energy expenditure Weight gain / Weight loss 2

Conclusions The most advanced tools for computer process description seem to be also the best tools for the description of biomolecular systems This intellectual economy validates the decades-long study of concurrency in computer science An essential foundation for the forthcoming “Virtual Cell Project”