DNA Storage 04/13/2020
66 Slides7.92 MB

DNA Storage 04/13/2020

Outlines DNA storage overall structure Encode/ECC code Basic operations: synthesis and sequencing Some existing works for DNA storage: DNA archive storage system OligoArchive for database system Puddle: A Dynamic, Error-Correcting, Full-Stack Microfluidics Platform

Basics Nucleotides: molecules that form the building blocks of DNA. Adenine (A) Thymine (T) Cytosine (C) Guanine (G), Naturally occurring DNA double helix with two strands of nucleotides DNA for data storage: oligonucleotide (oligo) a sequence of nucleotides Synthetic/Artifical DNA: synthesized using a chemical process that assembles the DNA one nucleotide at a time.
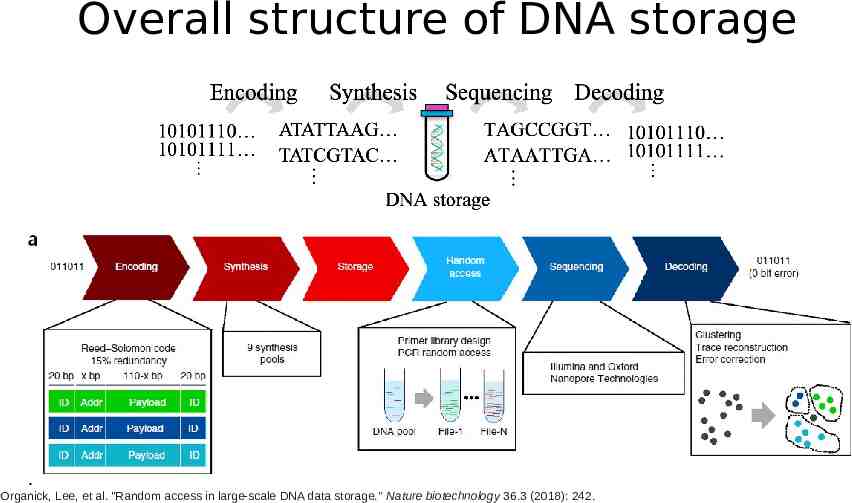
Overall structure of DNA storage Organick, Lee, et al. "Random access in large-scale DNA data storage." Nature biotechnology 36.3 (2018): 242.
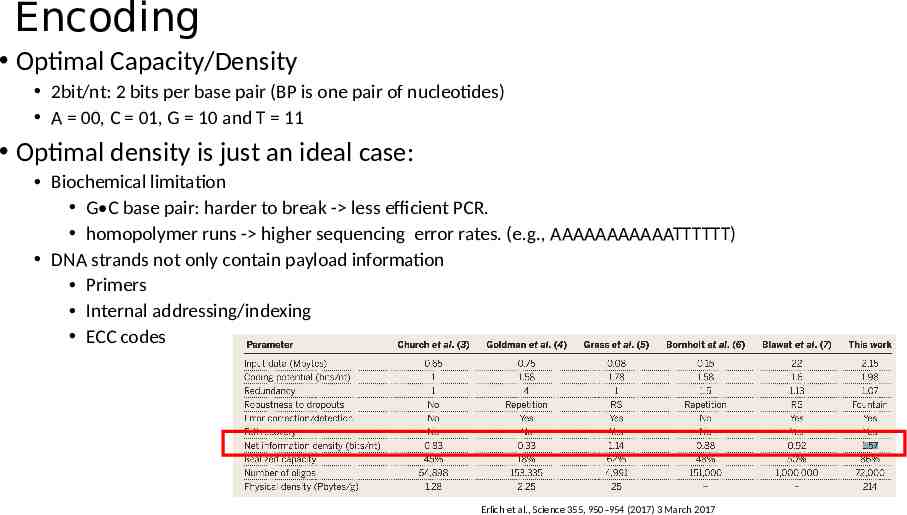
Encoding Optimal Capacity/Density 2bit/nt: 2 bits per base pair (BP is one pair of nucleotides) A 00, C 01, G 10 and T 11 Optimal density is just an ideal case: Biochemical limitation G C base pair: harder to break - less efficient PCR. homopolymer runs - higher sequencing error rates. (e.g., AAAAAAAAAAATTTTTT) DNA strands not only contain payload information Primers Internal addressing/indexing ECC codes Erlich et al., Science 355, 950–954 (2017) 3 March 2017

Encoding examples in DNA storage Early Works (inefficient error-prone ) Run number of 0 or 1. Morse code: (C dot, T dash, A word space and G letter space) [Church’12] (A, C 0 and T, G 1) George M. Church et al. Science 2012;337:1628
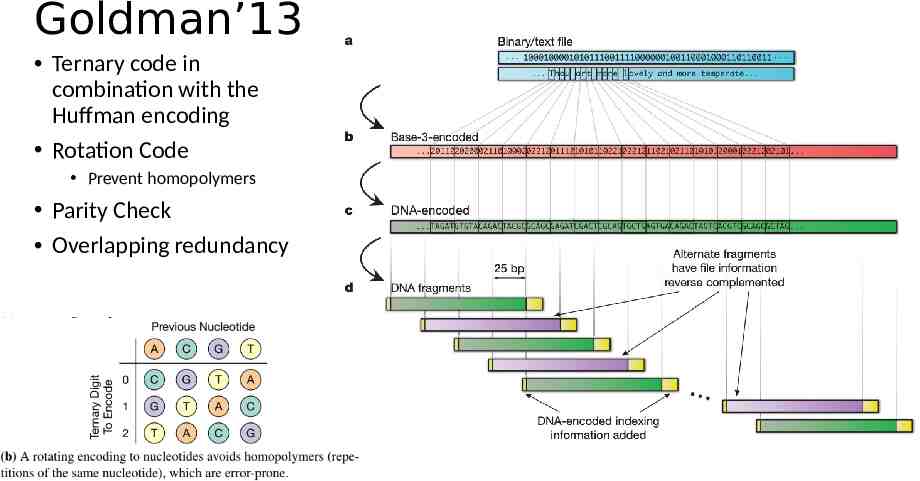
Goldman’13 Ternary code in combination with the Huffman encoding Rotation Code Prevent homopolymers Parity Check Overlapping redundancy
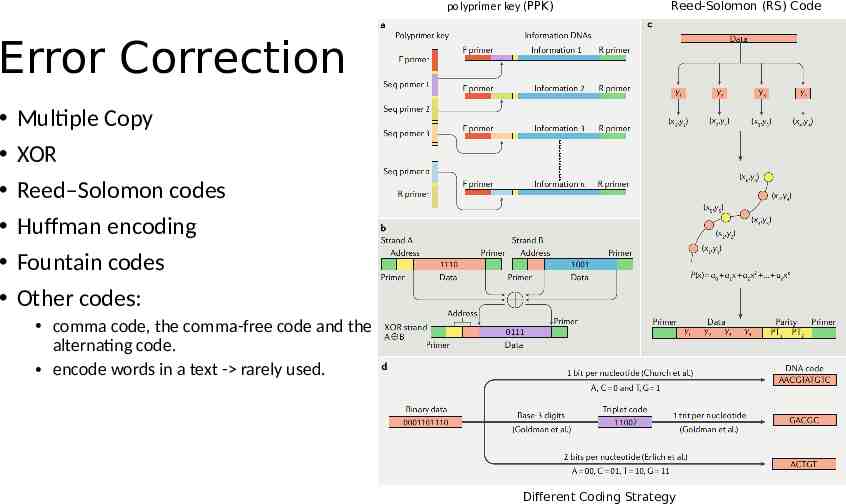
polyprimer key (PPK) Reed-Solomon (RS) Code Error Correction Multiple Copy XOR Reed–Solomon codes Huffman encoding Fountain codes Other codes: comma code, the comma-free code and the alternating code. encode words in a text - rarely used. Different Coding Strategy
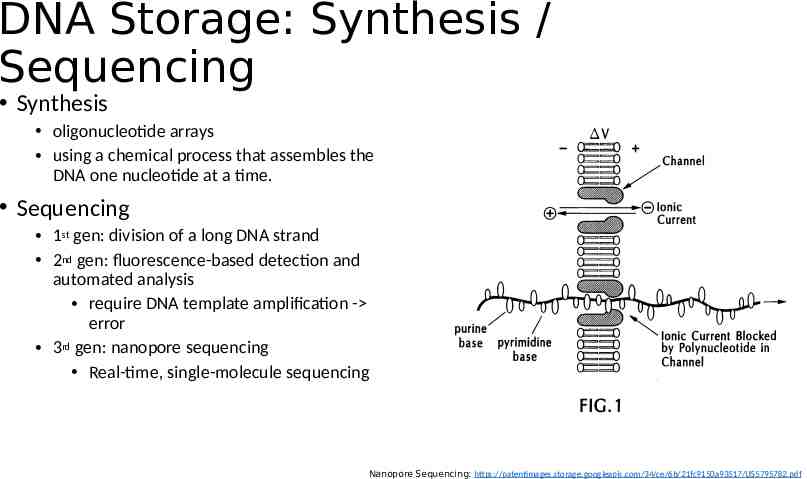
DNA Storage: Synthesis / Sequencing Synthesis oligonucleotide arrays using a chemical process that assembles the DNA one nucleotide at a time. Sequencing 1st gen: division of a long DNA strand 2nd gen: fluorescence-based detection and automated analysis require DNA template amplification - error 3rd gen: nanopore sequencing Real-time, single-molecule sequencing Nanopore Sequencing: https://patentimages.storage.googleapis.com/34/ce/6b/21fc9150a93517/US5795782.pdf
DNA Synthesis https://www.youtube.com/watch?time continue 3&v rD5uNAMbDaQ&feature emb title
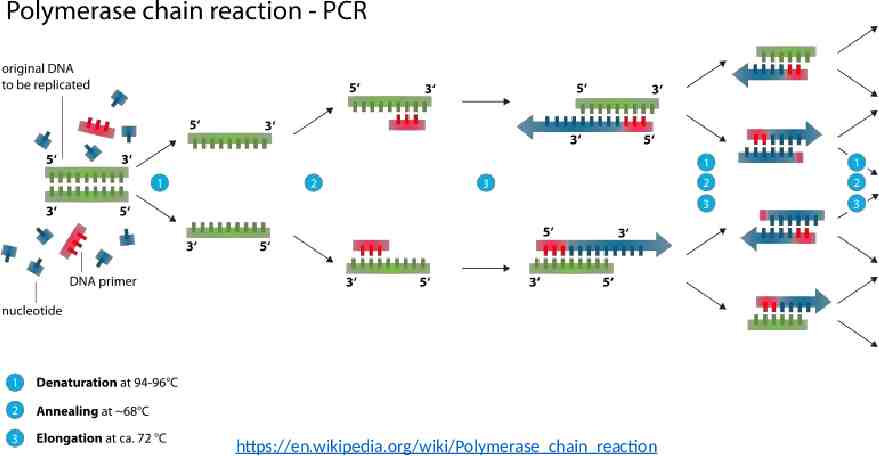
https://en.wikipedia.org/wiki/Polymerase chain reaction
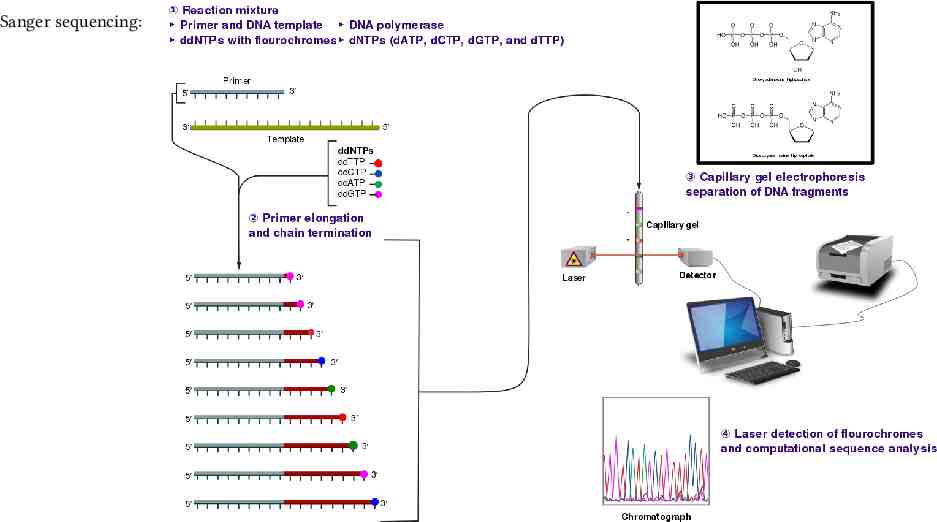
Sanger sequencing:

Limitations of DNA Synthesis/Sequencing Data layout and random access Max oligo length: ranges from a few hundred to a few thousand nucleotides at best. No logical addressing like block-based disk and tape. Address should be encoded along Synthesis & sequencing errors Synthesis: longer oligo more errors and truncated by products Sequencing: short-read - more substitution errors; long-read inserts or deletes spurious nucleotides. Error correction code is necessary

A DNA-Based Archival Storage System James Bornholt† Randolph Lopez† Douglas M. Carmean‡ Luis Ceze† Georg Seelig† Karin Strauss‡ † University of Washington ‡ Microsoft Research ASPLOS’16 Presented by: Fenggang Wu 7/12/2019

Summary Context: the exponential growth rate easily exceeds our ability to store it. Opportunity: DNA is extremely dense and long lasting. Problem: How to store and retrieve data based on DNA? Solution: DNA-based archival storage system Key-value store. Random access. Evaluations using wet lab experiment and simulation. Demonstrate feasibility, random access, and robustness.
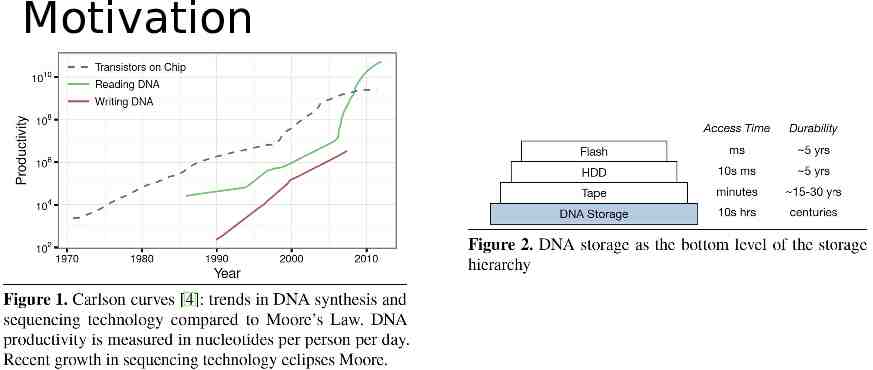
Motivation
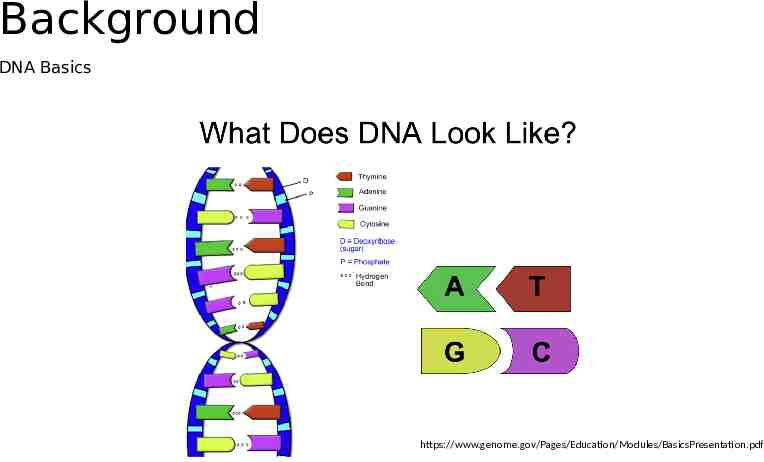
Background DNA Basics https://www.genome.gov/Pages/Education/Modules/BasicsPresentation.pdf

Background PCR: polymerase chain reaction PCR: a method for exponentially amplifying the concentration of selected sequences of DNA within a pool. Primers: The DNA sequencing primers are short synthetic strands that define the beginning and end of the region to be amplified.

Background DNA Synthesis Arbitrary single-strand DNA sequences can be synthesized chemically, nucleotide by nucleotide. Synthesizing error limits the size of the oligonucleotides ( 200 nucleotides). truncated byproducts Parallel synthesize [1]: synthesize 10 5 different oligonucleotides in parallel. [1] S. Kosuri and G. M. Church. Large-scale de novo DNA synthesis: technologies and applications. Nature Methods, 11:499–507, 2014.

Background DNA sequencing The DNA strand of interest serves as a template for PCR. Fluorescent nucleotides are used during this sequencing process. Read out the complement sequence optically. Read error. ( 1%)
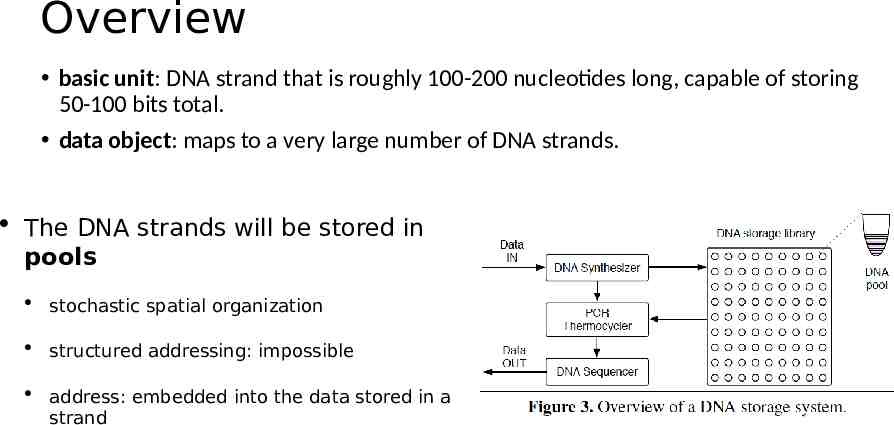
Overview basic unit: DNA strand that is roughly 100-200 nucleotides long, capable of storing 50-100 bits total. data object: maps to a very large number of DNA strands. The DNA strands will be stored in pools stochastic spatial organization structured addressing: impossible address: embedded into the data stored in a strand

Interface and Addressing Object Store: Put(key, value) / Get(key). Random access: mapping a key to a pair of PCR primers. write: primers are added to the strands read: those same primers are used in PCR to amplify only the strands with the desired keys. Separating the DNA strands into a collection of pools: Primers: reaction/limited number Error-prone.
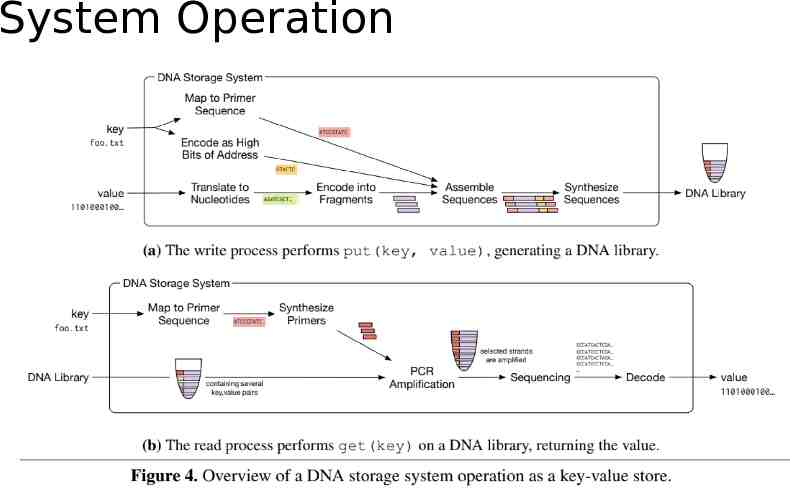
System Operation
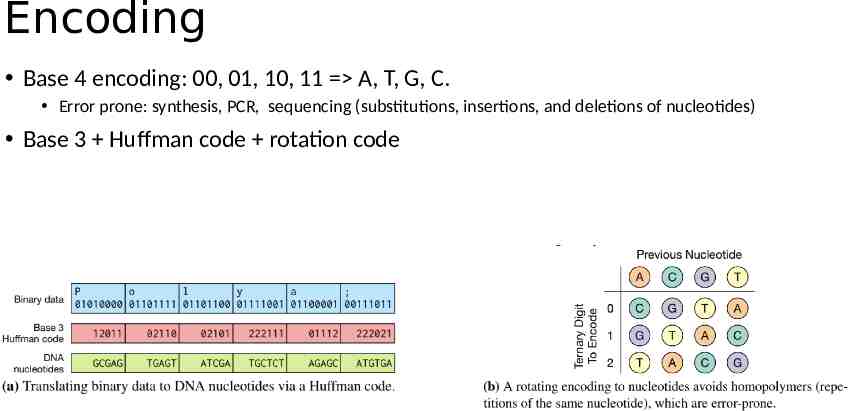
Encoding Base 4 encoding: 00, 01, 10, 11 A, T, G, C. Error prone: synthesis, PCR, sequencing (substitutions, insertions, and deletions of nucleotides) Base 3 Huffman code rotation code
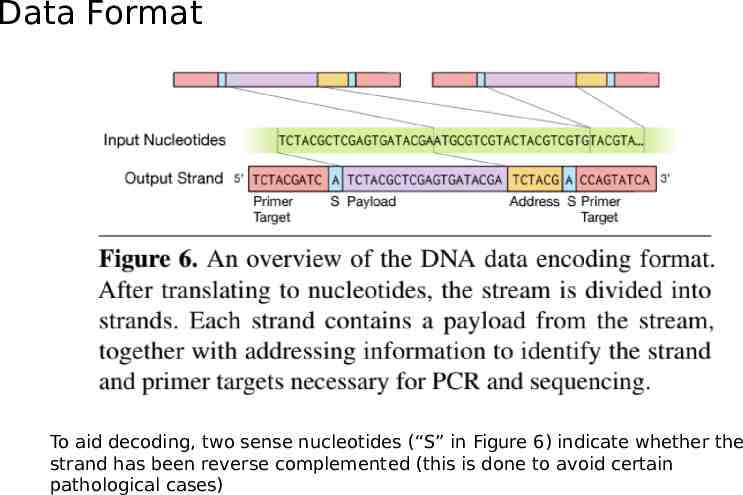
Data Format To aid decoding, two sense nucleotides (“S” in Figure 6) indicate whether the strand has been reverse complemented (this is done to avoid certain pathological cases)
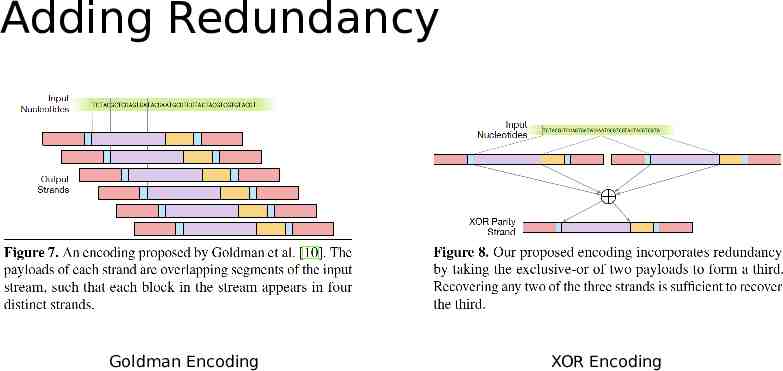
Adding Redundancy Goldman Encoding XOR Encoding

Other Factors Primer effectiveness Different error rate in the location of DNA strand Tunable redundancy

Evaluation Wet lab experiment Simulation
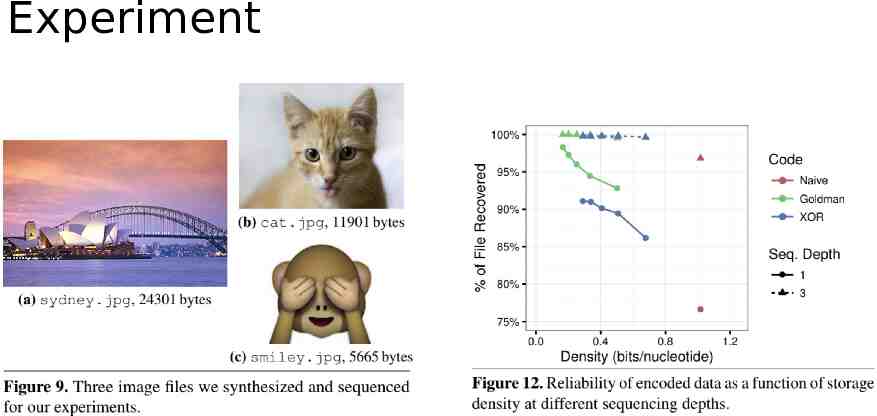
Experiment
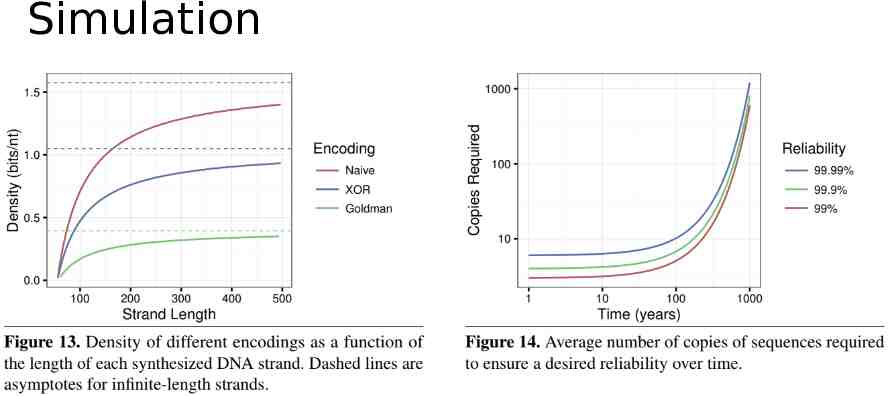
Simulation

Summary DNA-based storage has the potential to be the ultimate archival storage solution: it is extremely dense and durable. Background: Basics, Synthesize, PCR, Sequencing DNA archival system design: Overview, addressing (primer), data format (payload), encoding, redundancy. Evaluation (Experiment, Simulation) feasibility, random access, and robustness Conclusion: practicality. Time to borrow back from the biotechnology indusy.

Background

Limitations of DNA Synthesis/Sequencing Data layout and random access Max oligo length: ranges from a few hundred to a few thousand nucleotides at best. No logical addressing like block-based disk and tape. Address should be encoded along Synthesis & sequencing errors Synthesis: longer oligo more errors and truncated by products Sequencing: short-read - more substitution errors; long-read inserts or deletes spurious nucleotides. Error correction code is necessary

Limitations of DNA Synthesis/Sequencing Structural complexity Patterns amplifying both synthesis and sequencing errors homopolymer repeats (multiple consecutive occurrence of the same nucleotide) high GC content (higher Gs and Cs than As and Ts) Low-complexity regions - more error in sequencing same sequence of simple amino acids at similar positions. Encoder should avoid such problematic oligonucleotide sequences
Design

Overview Encoding/Decoding Synthesizing/Sequencing
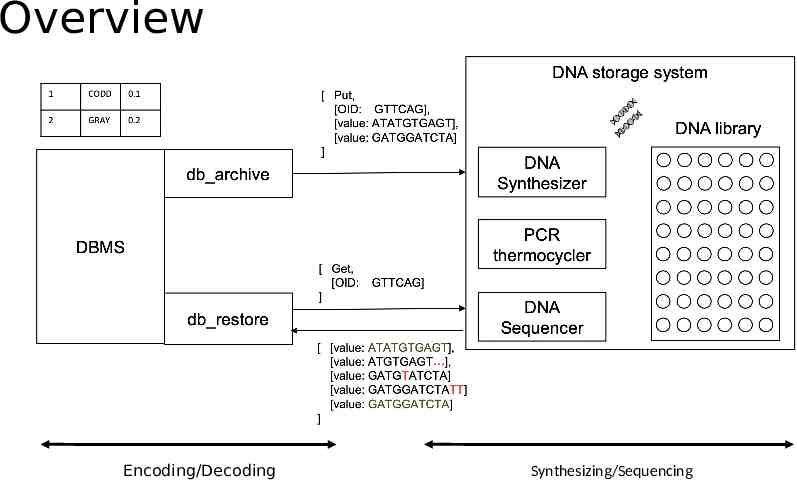
Design DNA Data Storage Query Processing Encoding Data Decoding Data Selection/Projection Join

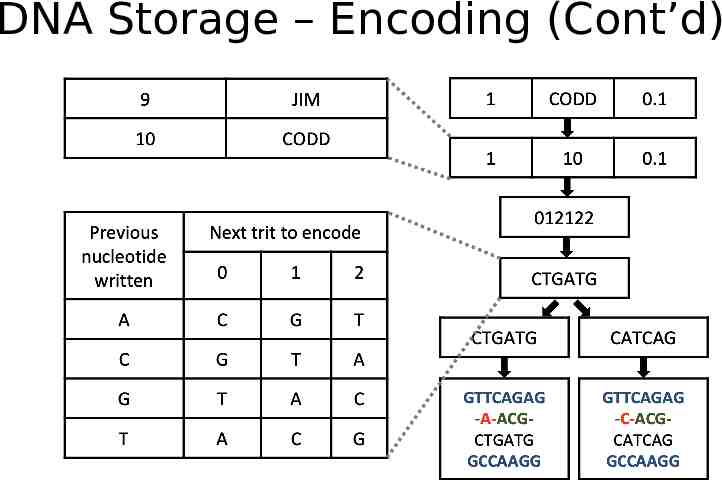
DNA Storage -- Encoding Extraction and preprocessing dictionary encoding: convert variable length string fields into fixed length integers DNA data representation Integers stream - ternary digit (Huffman code, base3) - Nucleotide (rotation code) Schema-aware encoding primary keys - primer. short records - pack into one oligo. long records, breaking by attributes and store in multiple oligos. Error correction metadata Detect: parity nucleotide; Correction: duplicating and reverse complementing oligo

DNA Storage – Encoding (Cont’d)
DNA Storage -- Decoding Merging the forward and reverse reads perform valid check to remove invalid oligos Translate back to data Nucleotides - trinary digit (base 3) - data Restore the records using the data

Query Processing DNA used for computation: excellent parallelism Hamiltonian path problem [1] or the strategic assignment problem [27]. See paper replicate logical gates using DNA polymerase [5] Neither provides new computational capacity, nor is particularly fast But, it can work on countless oligos in parallel Usage in Query Processing PCR DNA assembly methods
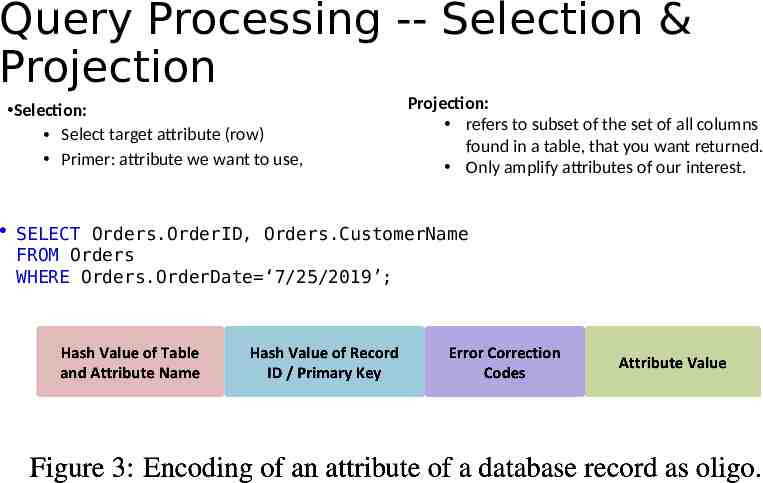
Query Processing -- Selection & Projection Selection: Select target attribute (row) Primer: attribute we want to use, Projection: refers to subset of the set of all columns found in a table, that you want returned. Only amplify attributes of our interest. SELECT Orders.OrderID, Orders.CustomerName FROM Orders WHERE Orders.OrderDate ‘7/25/2019’;
Query Processing -- Join SELECT Orders.OrderID, Customers.CustomerName FROM Orders INNER JOIN Customers ON Orders.CustomerID Customers.Cus tomerID WHERE Orders.OrderID 12345; CustomerID OrderID CustomerName CustomerID

Evaluation

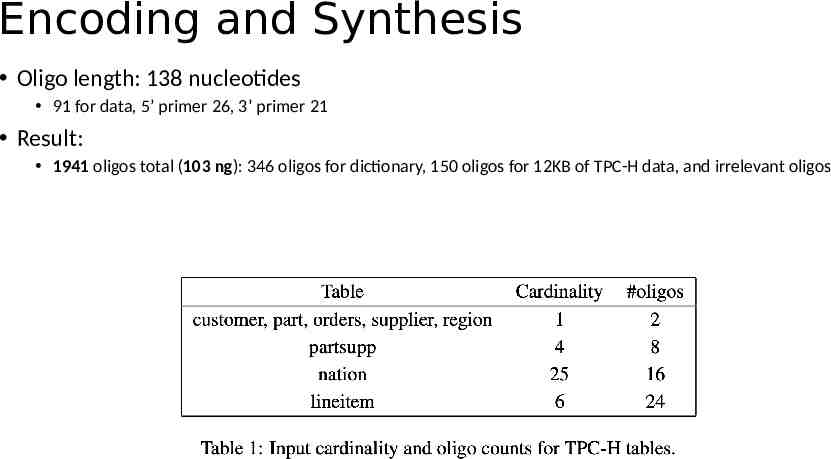
Setup PostgreSQL v10.3 pg oligo dump - file pg oligo restore - file Sequencing: Illumina NextSeq 500 platform TPC-H SF 10 4 dataset: 12KB, 44 records

Encoding and Synthesis Oligo length: 138 nucleotides 91 for data, 5’ primer 26, 3’ primer 21 Result: 1941 oligos total (103 ng): 346 oligos for dictionary, 150 oligos for 12KB of TPC-H data, and irrelevant oligos
Query Processing Selection: a single record is successfully selected. Join: one pair of matching oligos joins in a background of 105 irrelevant or nonmatching oligos.

Summary DNA is promising Storage Media: Density, durability OligoArchive: an architecture for using DNA as the archival tier of a relational DBMS Data Archival: Dump and restore Data Query: selection, projection, join

Extensions Not all SQL semantics are supported complex WHERE clause: non-equi selection complex join: non-equi-join multi-table join File system semantic: how to efficiently support modification? Scalability to large data sets Parallelism approximate string matching? Selection and projection using one oligo?
References DNA Synthesizing: https://www.youtube.com/watch?v rD5uNAMbDaQ PCR: https://www.youtube.com/watch?v 3XPAp6dgl14 DNA Sequencing: https://www.youtube.com/watch?v ONGdehkB8jU Gibson Assembly Cloning: https://www.youtube.com/watch?v tlVbf5fXhp4
Puddle: A Dynamic, ErrorCorrecting, Full-Stack Microfluidics Platform ASPLOS’2019 Presented by Bingzhe Li 2020/01/14

Lighting Talk https://www.youtube.com/watch?v uwiINEcYXLQ&list PL T9xA0eFRMdxHuQwkhLoa5No-wTSx3Tc&index 14&t 0s

Motivation Microfluidic device automates wetlab procedures by manipulating small chemical or biological samples. Current designs: Inflexible Error-prone Prohibitively expensive Difficult to program

Microfluidic Hardware (a) Channel-based: offer high precision and low cost at scale. (b) Liquid handling robots: more general, aiming to emulate a lab technician with robotic arms controlling pipettes (c) DMF: flexibility at small size and potentially at low cost called electrowetting on dielectric: activating electrodes in certain patterns can move, mix, or split droplets anywhere on the chip

Basic Controls in DMF devices Turning individual electrodes on or off To move a droplet from one location to another, a controller must activate the electrodes along that path in sequence DAG (directed acyclic graph issue)/routine issue in VLSI

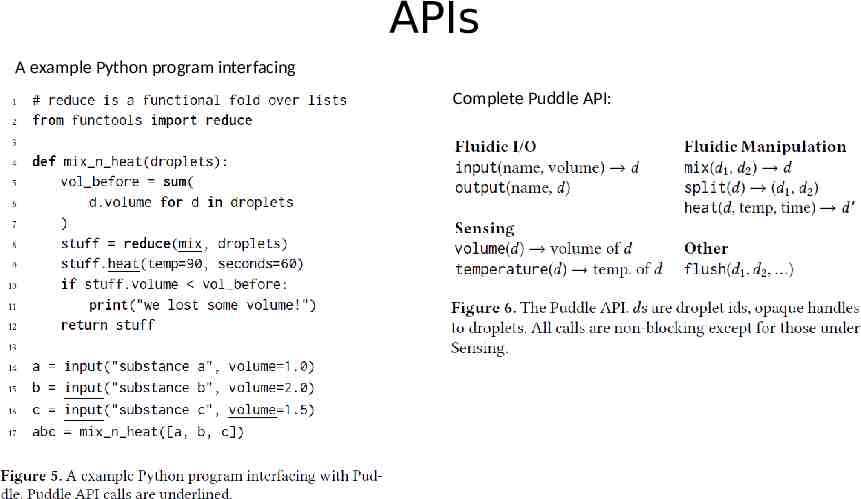
Dynamic Microfuidic Programming Vision of this paper: a microfluidics platform that can combine computation and fluidic manipulation in an unrestricted, high-level programming model a runtime system that provides a high-level API for microfluidic manipulations. APIs Handling errors Hardware implementation

APIs A example Python program interfacing Complete Puddle API:
Error Handling Two reasons cause API calls fail: Invalid arguments Using a consumed droplet id (not reuse droplets ids that have been consumed) Invalid arguments:

Implementation Three levels of the stack
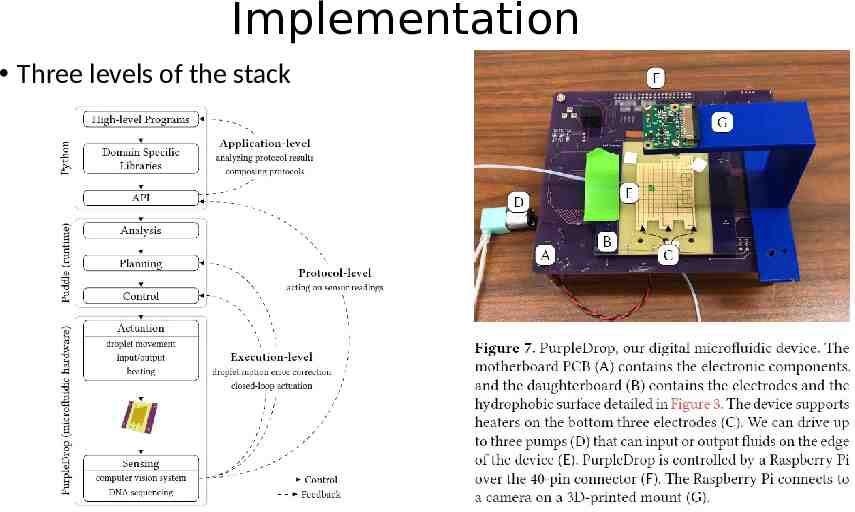
PurpleDrop DMF Device PurpleDrop, all together, the components cost on the order of 300, orders of magnitude less than most other microfluidic systems. DMF hardware: mother board contains the electrical components such as high-voltage controllers, shift registers, etc. daughterboard contains the electrodes that hold the droplets The daughterboard is removable, allowing different configurations of electrodes with the same motherboard. Electronic control: Raspberry Pi 3B The Raspberry Pi runs Linux and sports a quad-core 1.2 GHz ARMv7 processor as well as GPIO pins Peripherals: a heater, temperature sensor, and the ability to do both input and output of droplets. Input and output are driven by small peristaltic pumps which carry droplets to/from test tube reservoirs or other devices

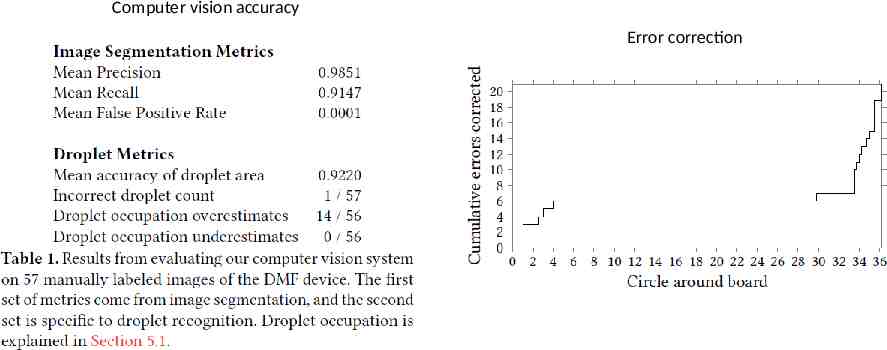
Planning and Execution The core of planning is placement and routing Allocation constraints: A mix requests a rectangle slightly larger than the droplet to move and agitate droplets Place constraints (heater position) Execution, Monitoring, and Rollback Execution: activating the electrodes Monitoring: computer vision system to check the states of droplets on the board A rollback consists of deleting the record and replanning all commands which have not been completed.

Evaluation Error system Computer vision Error correction PCR and Thermocycling DNA sequencing
Computer vision accuracy Error correction
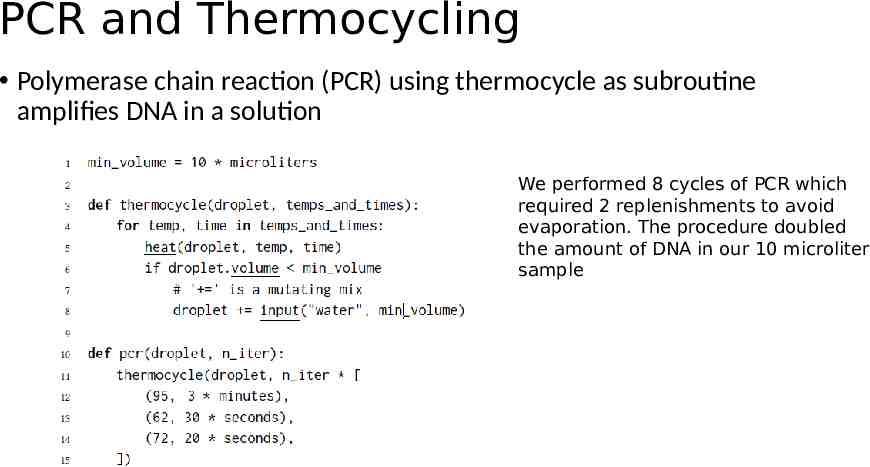
PCR and Thermocycling Polymerase chain reaction (PCR) using thermocycle as subroutine amplifies DNA in a solution We performed 8 cycles of PCR which required 2 replenishments to avoid evaporation. The procedure doubled the amount of DNA in our 10 microliter sample
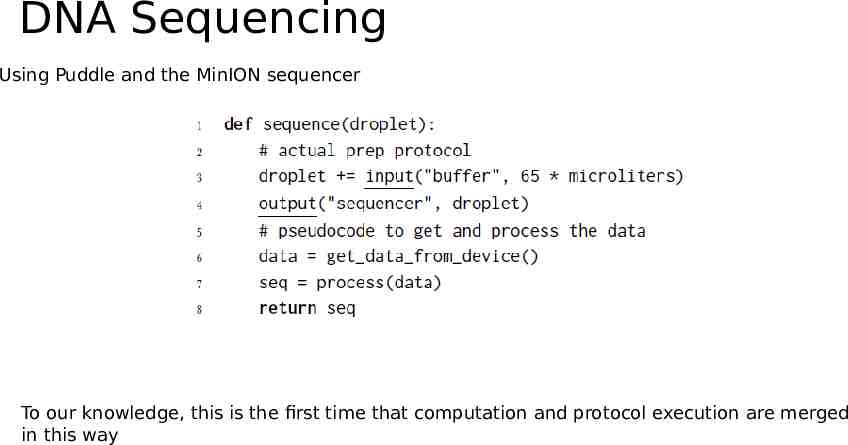
DNA Sequencing Using Puddle and the MinION sequencer To our knowledge, this is the first time that computation and protocol execution are merged in this way