APPLICATIONS OF MOLECULAR BIOLOGY TECHNIQUES TO MEDICAL MICROBIOLOGY
14 Slides39.00 KB

APPLICATIONS OF MOLECULAR BIOLOGY TECHNIQUES TO MEDICAL MICROBIOLOGY

Two principle molecular techniques used in detection of microorganisms 1- Nucleic acid hybridization( Southern Blotting) 2- Polymerase chain reaction (PCR)

Polymerase chain reaction The benefits of PCR based diagnostic testing: Rapid diagnosis Detection Same day result High accuracy, high specifity,high sensitivity

The role of PCR in diagnosis of infectious diseases Fastidious and slow growing microorganism Detect antimicrobial resistance Detect microorganism cannt be cultivated Measurment value of viral load
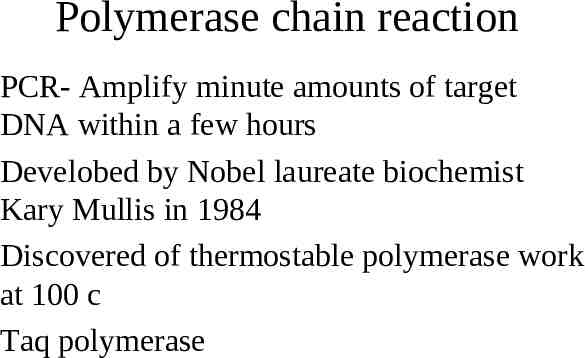
Polymerase chain reaction PCR- Amplify minute amounts of target DNA within a few hours Develobed by Nobel laureate biochemist Kary Mullis in 1984 Discovered of thermostable polymerase work at 100 c Taq polymerase

PCR methodology Materials Target DNA Taq polymerase Four DNA nucleotides Tow primerS Reaction buffer Temperature cycles

PCR methods 1- Denaturation: doble stranded DNA eprated into two single strands 90-95c 2- Cooling at 30-60c 3- Annealing: Primers attached complementary region of target DNA 4- Heating at 70c 5- Extention :The primers extended to form a new strand of DNA
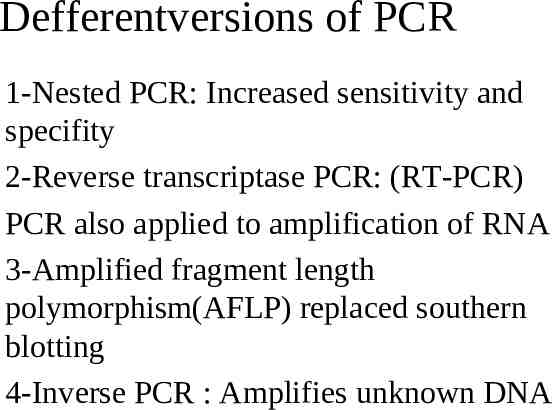
Defferentversions of PCR 1-Nested PCR: Increased sensitivity and specifity 2-Reverse transcriptase PCR: (RT-PCR) PCR also applied to amplification of RNA 3-Amplified fragment length polymorphism(AFLP) replaced southern blotting 4-Inverse PCR : Amplifies unknown DNA
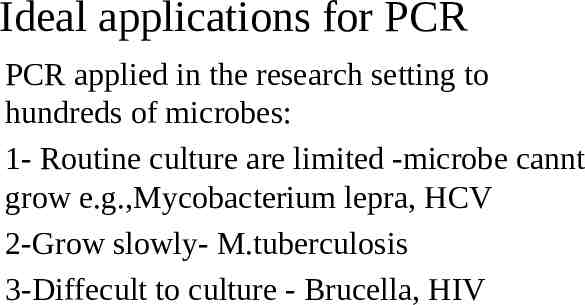
Ideal applications for PCR PCR applied in the research setting to hundreds of microbes: 1- Routine culture are limited -microbe cannt grow e.g.,Mycobacterium lepra, HCV 2-Grow slowly- M.tuberculosis 3-Diffecult to culture - Brucella, HIV

Diagnosis of infectious diseases Examples of infection agents that detected by PCR: Chlamydia trachomatis C. pneumonia Mycobacterium tuberculosis Mycoblasma pneumoniae Neisseria gonorrhoeae Herbes simplex virus
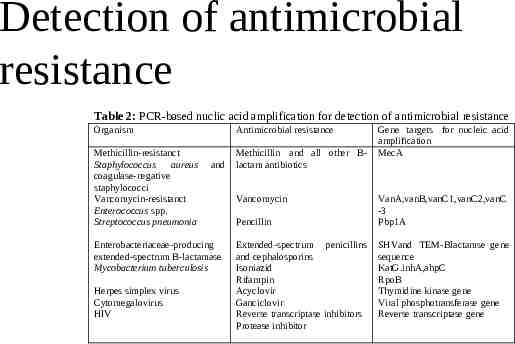
Detection of antimicrobial resistance Table 2: PCR-based nuclic acid amplification for detection of antimicrobial resistance Organism Methicillin-resistanct Staphylococcus aureus coagulase-negative staphylococci Vancomycin-resistanct Enterococcus spp. Streptococcus pneumonia Antimicrobial resistance and Enterobacteriaceae-producing extended-spectrum B-lactamase Mycobacterium tuberculosis Herpes simplex virus Cytomegalovirus HIV Methicillin and all other Blactam antibiotics Vancomycin Pencillin Extended-spectrum penicillins and cephalosporins Isoniazid Rifampin Acyclovir Ganciclovir Reverse transcriptase inhibitors Protease inhibitor Gene targets for nucleic acid amplification MecA VanA,vanB,vanC1,vanC2,vanC -3 Pbp1A SHVand TEM-Blactamse gene sequence KatG.inhA,ahpC RpoB Thymidine kinase gene Viral phosphotransferase gene Reverse transcriptase gene

Southern blotting Named after Edward M. Southern whodeveloped this procedure in 1975 Allow investigators to detrmine the molecular weight of a restriction fragment To measure relative amounts in different samples To locate a particular sequence of DNA within a complex mixture

Southern blotting methodology 1- Digest DNA with appropriate restriction enzyme 2- Run digest on a agrose gel 3- Denaturate the DNA 4- Transfer the denaturated DNA to the membrane 5- Add labeled probe to the membrane 6-Detection

Just a few application of southern blotting To look for or to confirm the presence of a gene often in conjugation with PCR To test for the presence of a specific allel of a gene To aid in restriction fragment analyses RFLP






