Transposition Evidence Mechanisms: DNA-mediated RNA-mediated
31 Slides1.31 MB
Transposition Evidence Mechanisms: DNA-mediated RNA-mediated
Transposable elements Mobile genetic elements - they move from one location in the genome to another Found in all organisms (so far studied) Effects: – Insertion near or within a gene can inactivate or activate the target gene. – Cause deletions, inversions, and translocations of DNA – Lead to chromosome breaks
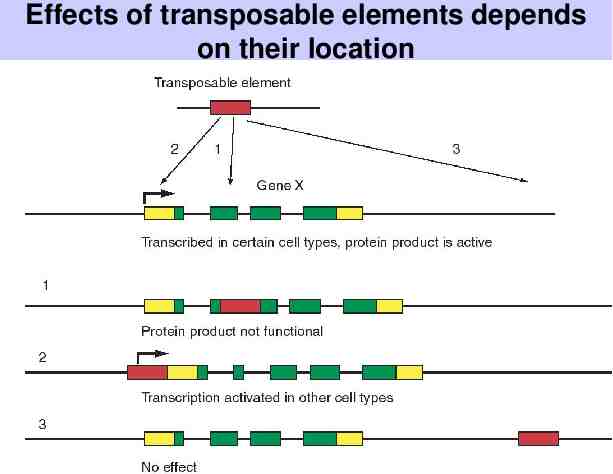
Effects of transposable elements depends on their location
Observations of B. McClintock (1930’s-1950’s) Certain crosses in maize resulted in large numbers of mutable loci. – The frequency of change at those loci is much higher than normally observed. Studies of these plants revealed a genetic element called “Dissociation” or Ds on the short arm of chromosome 9. Chromosome breaks occurred at the Ds locus, which could be observed cytologically – i.e. by looking at chromosome spreads from individual cells, e.g. sporocytes. Frequency and timing of these breaks is controlled by another locus, called “Activator” or Ac.
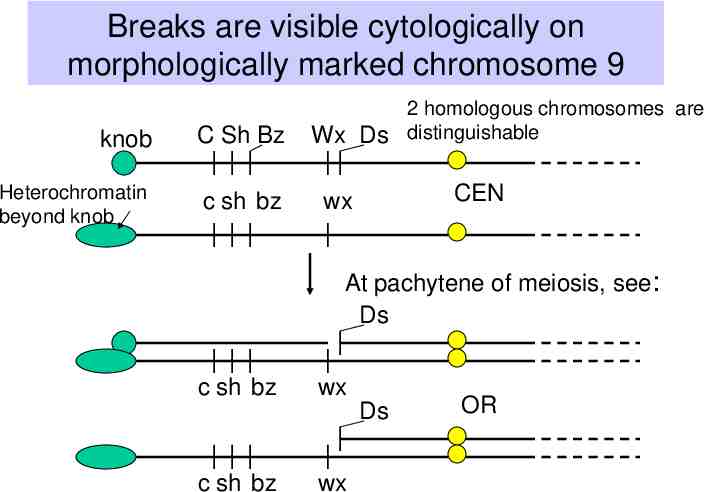
Breaks are visible cytologically on morphologically marked chromosome 9 knob C Sh Bz Heterochromatin beyond knob c sh bz 2 homologous chromosomes are Wx Ds distinguishable CEN wx At pachytene of meiosis, see: Ds c sh bz wx c sh bz wx Ds OR
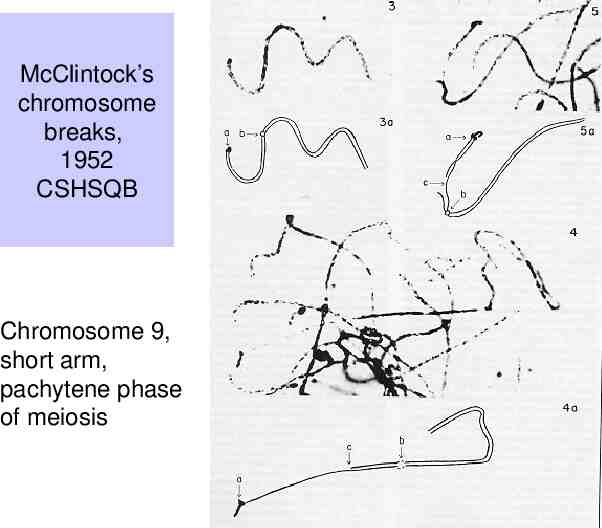
McClintock’s chromosome breaks, 1952 CSHSQB Chromosome 9, short arm, pachytene phase of meiosis
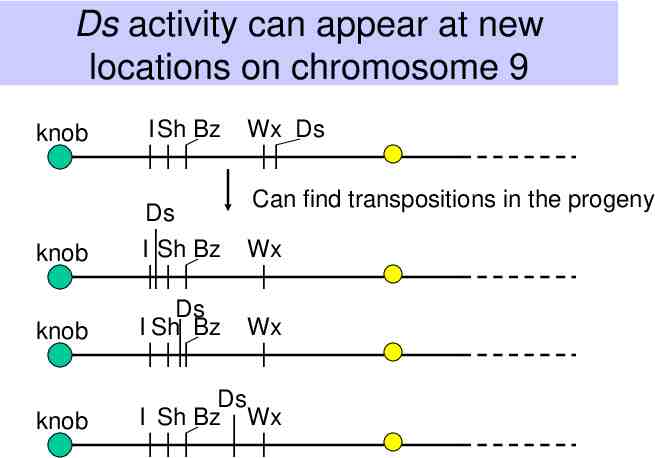
Ds activity can appear at new locations on chromosome 9 knob I Sh Bz Wx Ds Can find transpositions in the progeny knob Ds I Sh Bz Wx knob Ds I Sh Bz Wx knob Ds I Sh Bz Wx
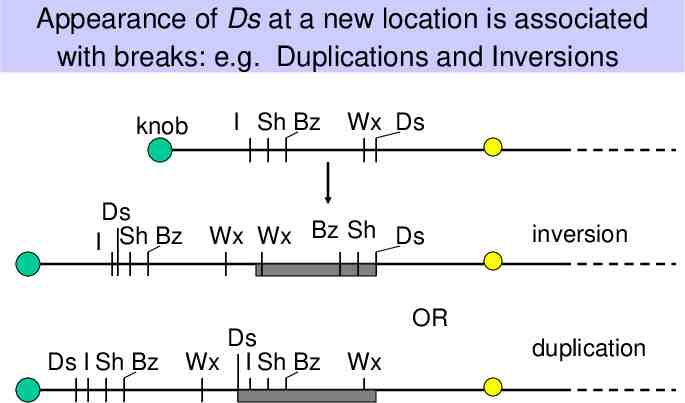
Appearance of Ds at a new location is associated with breaks: e.g. Duplications and Inversions knob Ds I Sh Bz Ds I Sh Bz I Sh Bz Wx Ds Wx Wx Bz Sh Ds Ds Wx I Sh Bz inversion OR Wx duplication
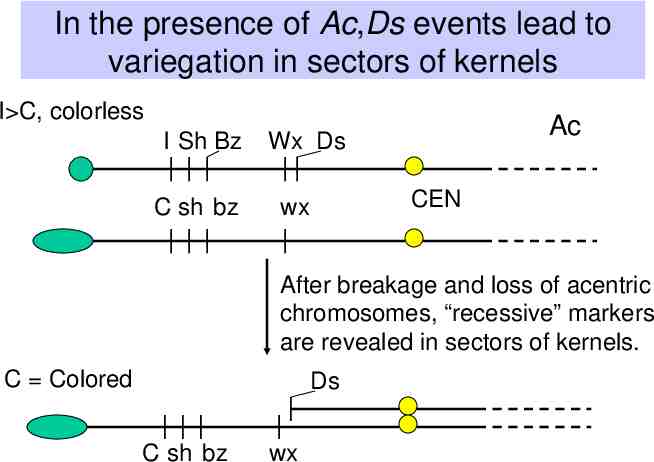
In the presence of Ac,Ds events lead to variegation in sectors of kernels I C, colorless I Sh Bz C sh bz Ac Wx Ds CEN wx After breakage and loss of acentric chromosomes, “recessive” markers are revealed in sectors of kernels. C Colored Ds C sh bz wx
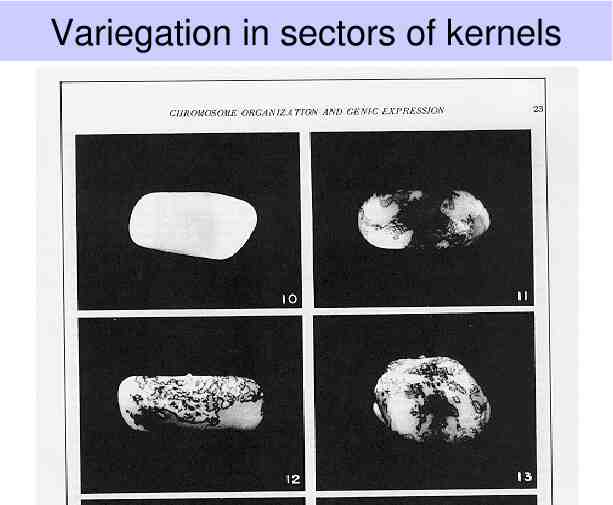
Variegation in sectors of kernels
Variegation in wild flox

Mechanisms of Transposition
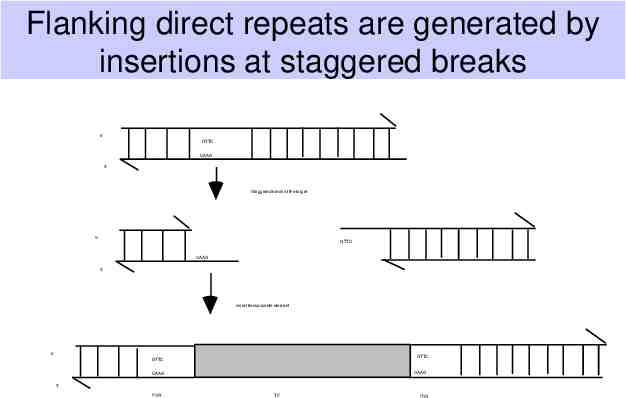
Flanking direct repeats are generated by insertions at staggered breaks Flanking direct repeats are generated by insertions at staggered breaks 5' GTTC CAAG 3' Staggered break at the target 5' GTTC CAAG 3' Insert transposable element 5' GTTC GTTC CAAG CAAG 3' FDR TE FDR
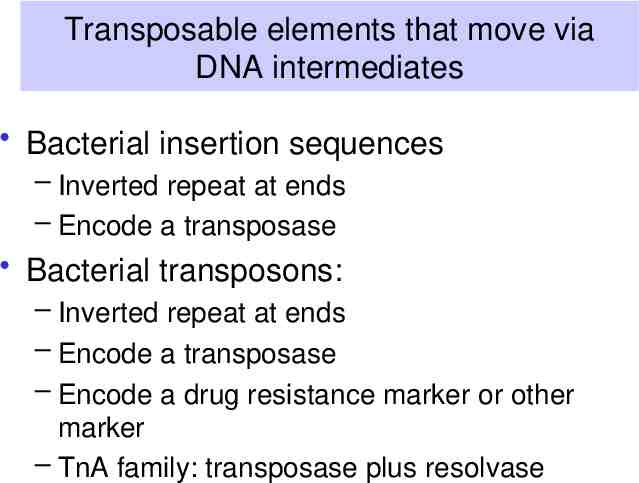
Transposable elements that move via DNA intermediates Bacterial insertion sequences – Inverted repeat at ends – Encode a transposase Bacterial transposons: – Inverted repeat at ends – Encode a transposase – Encode a drug resistance marker or other marker – TnA family: transposase plus resolvase
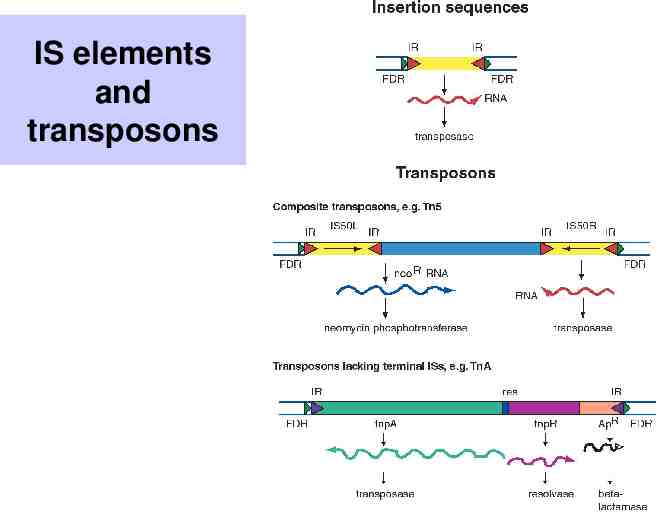
IS elements and transposons

Ac/Ds transposons in maize Ac is autonomous – Inverted repeats – Encodes a transposase Ds is nonautonomous – Inverted repeats – Transposase gene is defective because of deletions in coding region
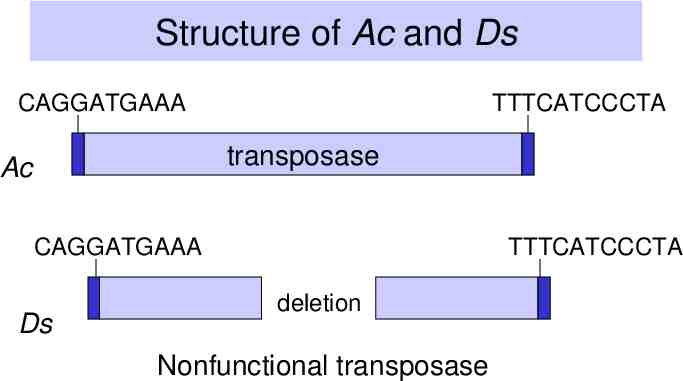
Structure of Ac and Ds CAGGATGAAA TTTCATCCCTA transposase Ac CAGGATGAAA Ds TTTCATCCCTA deletion Nonfunctional transposase
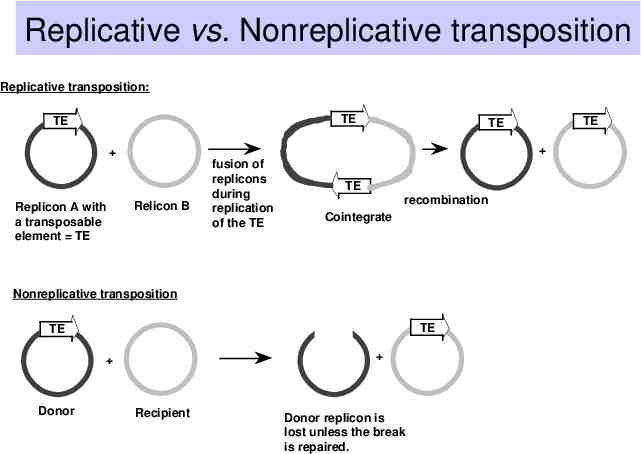
Replicative vs. Nonreplicative transposition Replicative transposition: TE TE Replicon A with a transposable element TE Relicon B fusion of replicons during replication of the TE TE recombination Cointegrate Nonreplicative transposition TE TE Donor TE TE Recipient Donor replicon is lost unless the break is repaired.
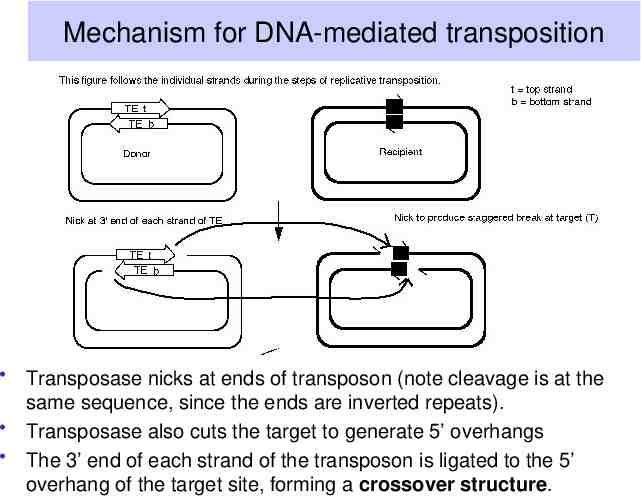
Mechanism for DNA-mediated transposition Transposase nicks at ends of transposon (note cleavage is at the same sequence, since the ends are inverted repeats). Transposase also cuts the target to generate 5’ overhangs The 3’ end of each strand of the transposon is ligated to the 5’ overhang of the target site, forming a crossover structure.
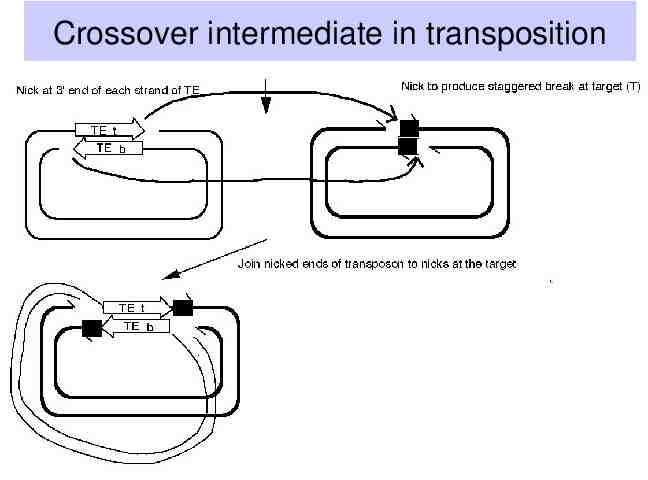
Crossover intermediate in transposition
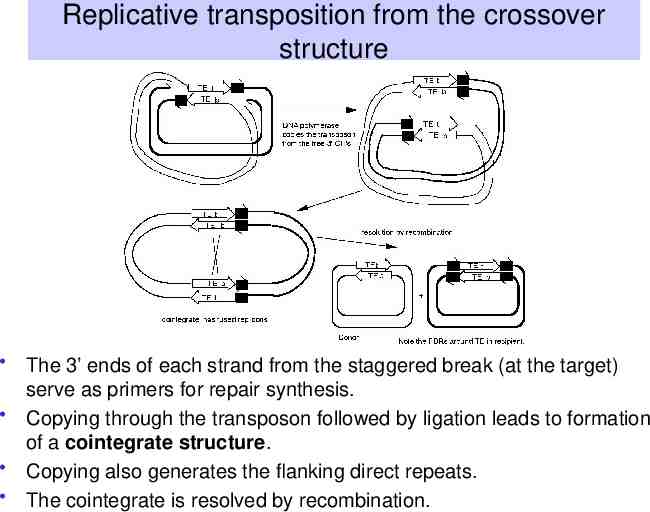
Replicative transposition from the crossover structure The 3’ ends of each strand from the staggered break (at the target) serve as primers for repair synthesis. Copying through the transposon followed by ligation leads to formation of a cointegrate structure. Copying also generates the flanking direct repeats. The cointegrate is resolved by recombination.
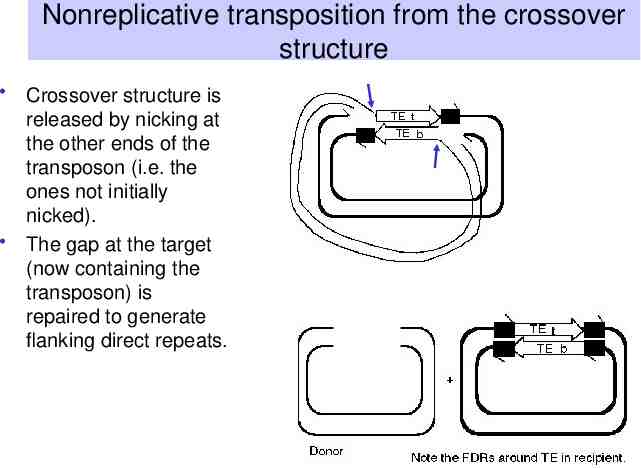
Nonreplicative transposition from the crossover structure Crossover structure is released by nicking at the other ends of the transposon (i.e. the ones not initially nicked). The gap at the target (now containing the transposon) is repaired to generate flanking direct repeats.
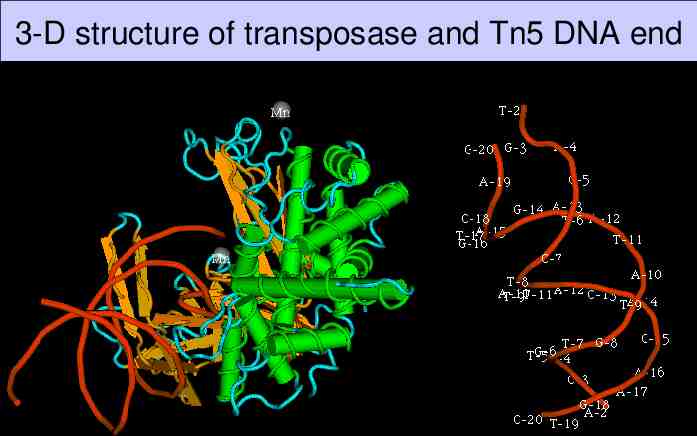
3-D structure of transposase and Tn5 DNA end
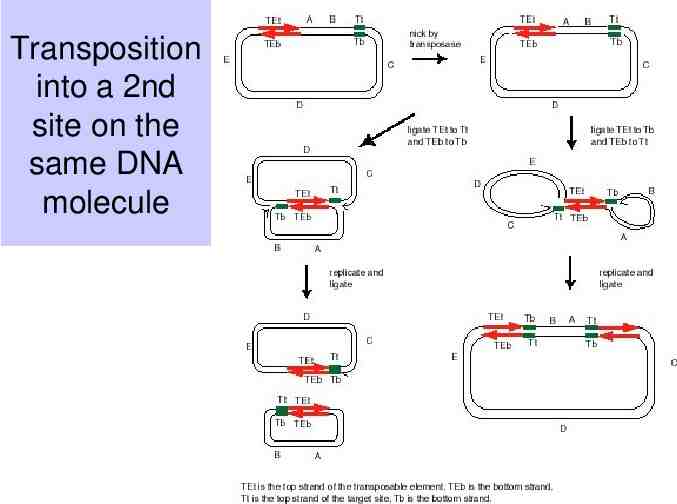
Transposition into a 2nd site on the same DNA molecule
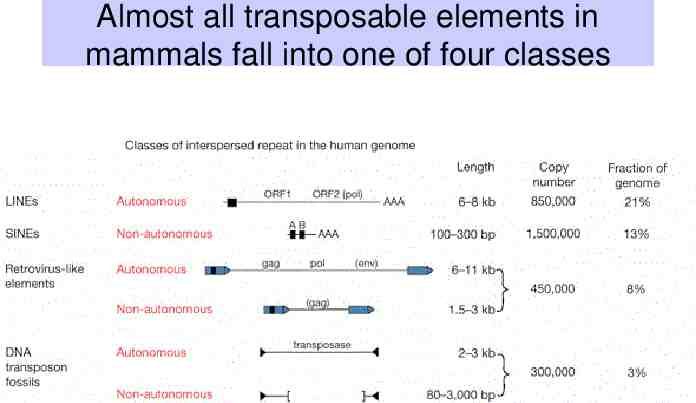
Almost all transposable elements in mammals fall into one of four classes
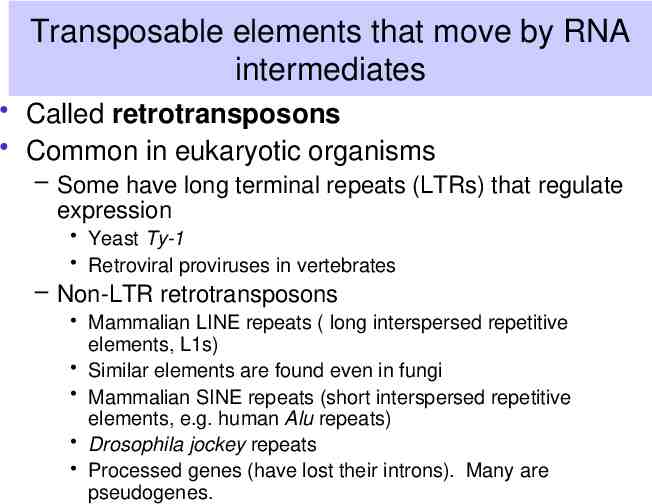
Transposable elements that move by RNA intermediates Called retrotransposons Common in eukaryotic organisms – Some have long terminal repeats (LTRs) that regulate expression Yeast Ty-1 Retroviral proviruses in vertebrates – Non-LTR retrotransposons Mammalian LINE repeats ( long interspersed repetitive elements, L1s) Similar elements are found even in fungi Mammalian SINE repeats (short interspersed repetitive elements, e.g. human Alu repeats) Drosophila jockey repeats Processed genes (have lost their introns). Many are pseudogenes.
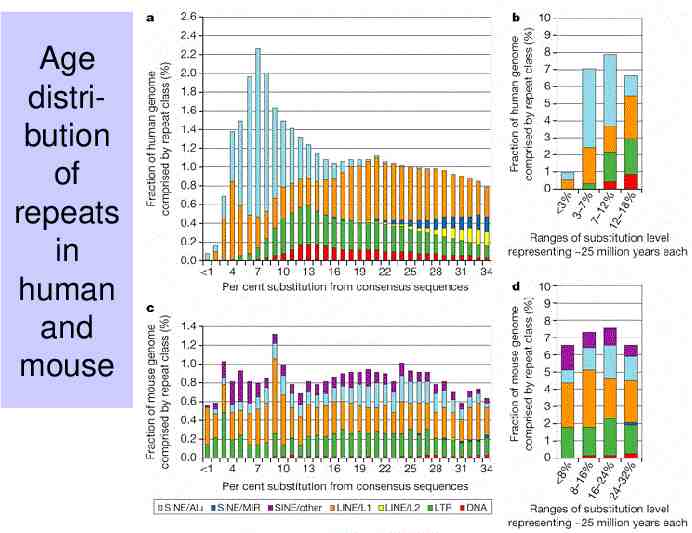
Age distribution of repeats in human and mouse
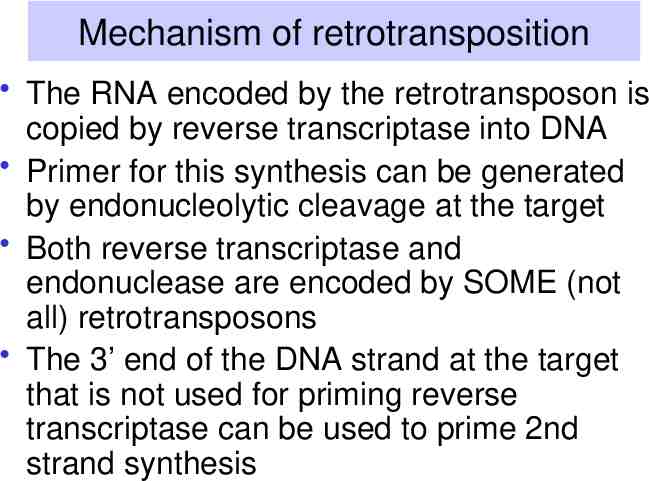
Mechanism of retrotransposition The RNA encoded by the retrotransposon is copied by reverse transcriptase into DNA Primer for this synthesis can be generated by endonucleolytic cleavage at the target Both reverse transcriptase and endonuclease are encoded by SOME (not all) retrotransposons The 3’ end of the DNA strand at the target that is not used for priming reverse transcriptase can be used to prime 2nd strand synthesis
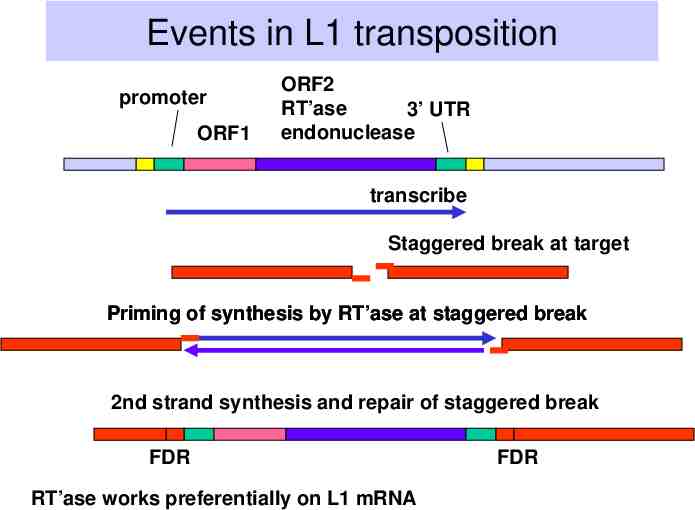
Events in L1 transposition promoter ORF1 ORF2 RT’ase 3’ UTR endonuclease transcribe Staggered break at target Priming of synthesis by RT’ase at staggered break 2nd strand synthesis and repair of staggered break FDR RT’ase works preferentially on L1 mRNA FDR

Recombination between two nearly identical sequences (e.g transposons) will lead to rearrangements Deletion if the repeats are in the same orientation Inversion if the repeats are in the opposite orientation
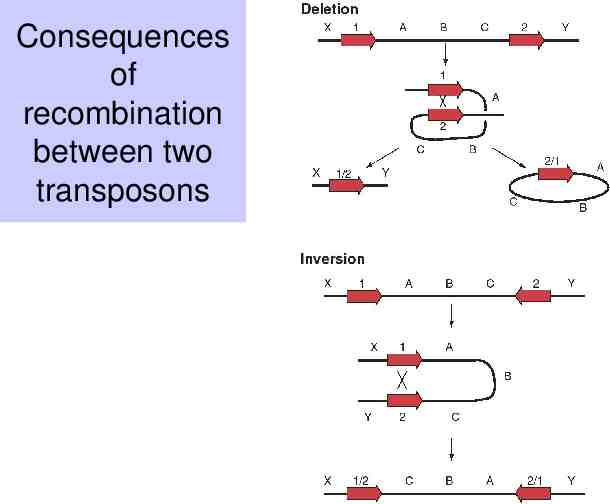
Consequences of recombination between two transposons






