Welcome! Mass spectrometry meets cheminformatics Tobias Kind and
25 Slides3.56 MB
Welcome! Mass spectrometry meets cheminformatics Tobias Kind and Julie Leary UC Davis Course 2: Mass spectral and molecular data handling Class website: CHE 241 - Spring 2008 - CRN 16583 Slides: http://fiehnlab.ucdavis.edu/staff/kind/Teaching/ PPT is hyperlinked – please change to Slide Show Mode 1
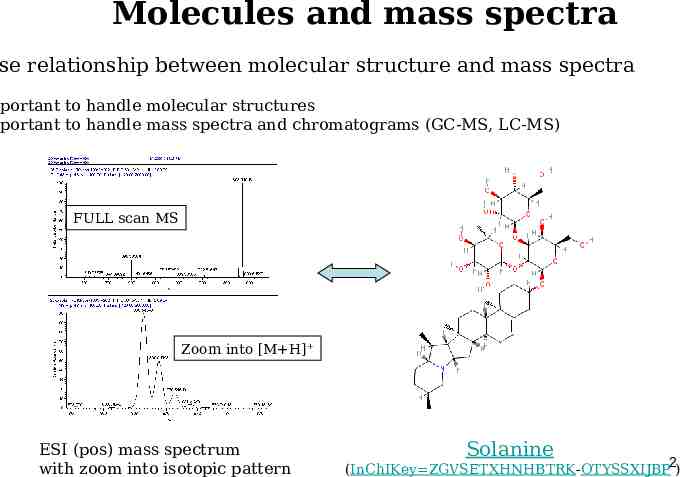
Molecules and mass spectra se relationship between molecular structure and mass spectra mportant to handle molecular structures mportant to handle mass spectra and chromatograms (GC-MS, LC-MS) FULL scan MS Zoom into [M H] ESI (pos) mass spectrum with zoom into isotopic pattern Solanine (InChIKey ZGVSETXHNHBTRK-OTYSSXIJBP2)
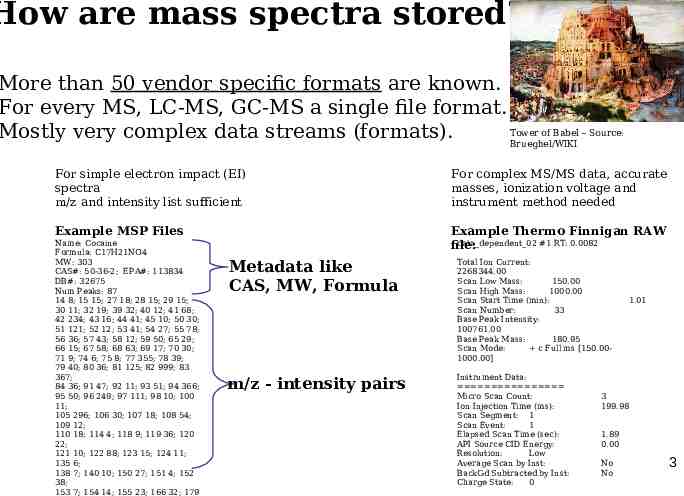
How are mass spectra stored? More than 50 vendor specific formats are known. For every MS, LC-MS, GC-MS a single file format. Tower of Babel – Source: Mostly very complex data streams (formats). Brueghel/WIKI For simple electron impact (EI) spectra m/z and intensity list sufficient For complex MS/MS data, accurate masses, ionization voltage and instrument method needed Example MSP Files Example Thermo Finnigan RAW data dependent 02 #1 RT: 0.0082 file: Name: Cocaine Formula: C17H21NO4 MW: 303 CAS#: 50-36-2; EPA#: 113834 DB#: 32675 Num Peaks: 87 14 8; 15 15; 27 18; 28 15; 29 15; 30 11; 32 19; 39 32; 40 12; 41 68; 42 234; 43 16; 44 41; 45 10; 50 30; 51 121; 52 12; 53 41; 54 27; 55 78; 56 36; 57 43; 58 12; 59 50; 65 29; 66 15; 67 58; 68 63; 69 17; 70 30; 71 9; 74 6; 75 8; 77 355; 78 39; 79 40; 80 36; 81 125; 82 999; 83 367; 84 36; 91 47; 92 11; 93 51; 94 366; 95 50; 96 249; 97 111; 98 10; 100 11; 105 296; 106 30; 107 18; 108 54; 109 12; 110 18; 114 4; 118 9; 119 36; 120 22; 121 10; 122 88; 123 15; 124 11; 135 6; 138 7; 140 10; 150 27; 151 4; 152 38; 153 7; 154 14; 155 23; 166 32; 179 Metadata like CAS, MW, Formula m/z - intensity pairs Total Ion Current: 2268344.00 Scan Low Mass: 150.00 Scan High Mass: 1000.00 Scan Start Time (min): Scan Number: 33 Base Peak Intensity: 100761.00 Base Peak Mass: 180.95 Scan Mode: c Full ms [150.001000.00] Instrument Data: Micro Scan Count: Ion Injection Time (ms): Scan Segment: 1 Scan Event: 1 Elapsed Scan Time (sec): API Source CID Energy: Resolution: Low Average Scan by Inst: BackGd Subtracted by Inst: Charge State: 0 1.01 3 199.98 1.89 0.00 No No 3
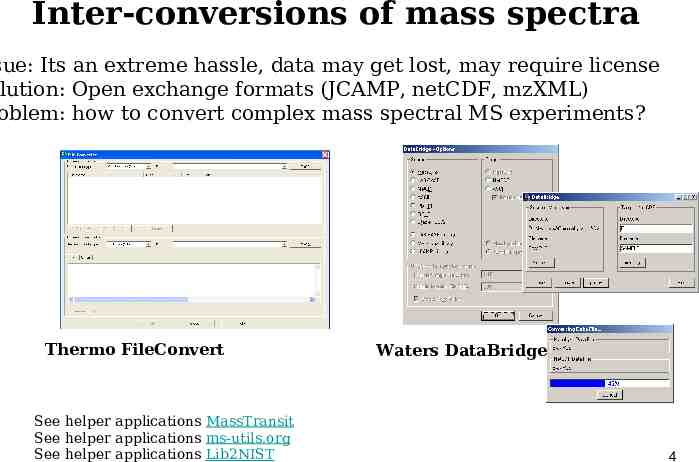
Inter-conversions of mass spectra sue: Its an extreme hassle, data may get lost, may require license lution: Open exchange formats (JCAMP, netCDF, mzXML) oblem: how to convert complex mass spectral MS experiments? Thermo FileConvert See helper applications MassTransit See helper applications ms-utils.org See helper applications Lib2NIST Waters DataBridge 4
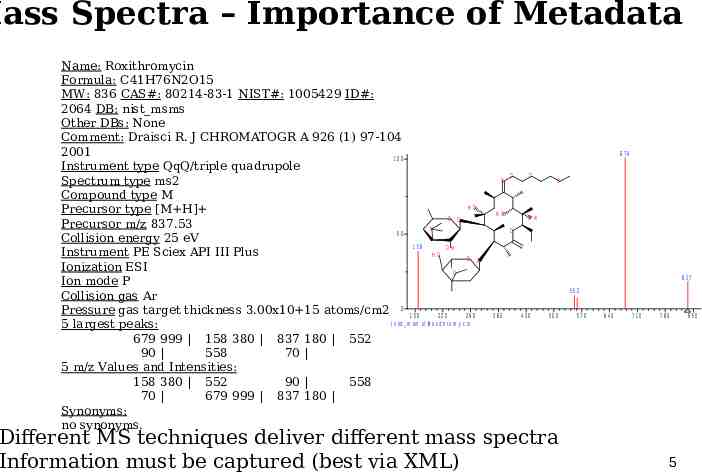
Mass Spectra – Importance of Metadata Name: Roxithromycin Formula: C41H76N2O15 MW: 836 CAS#: 80214-83-1 NIST#: 1005429 ID#: 2064 DB: nist msms Other DBs: None Comment: Draisci R. J CHROMATOGR A 926 (1) 97-104 2001 100 Instrument type QqQ/triple quadrupole Spectrum type ms2 Compound type M HO Precursor type [M H] O O Precursor m/z 837.53 N 50 Collision energy 25 eV 158 O H Instrument PE Sciex API III Plus HO O O Ionization ESI O Ion mode P Collision gas Ar Pressure gas target thickness 3.00x10 15 atoms/cm2 0 1 5 0 220 290 ( n is t m s m s ) R o x it h r o m y c in 5 largest peaks: 679 999 158 380 837 180 552 90 558 70 5 m/z Values and Intensities: 158 380 552 90 558 70 679 999 837 180 Synonyms: no synonyms. 679 N O O HO O O H O O 837 552 360 430 500 Different MS techniques deliver different mass spectra Information must be captured (best via XML) 570 640 710 780 850 5
pen Exchange formats for mass spectra hy? You’re in a successful lab using multiple vendor mass spectrometers hy? You want to share and receive mass spectra from colleagues. hy? Future grants will require depositing of mass spectra in repositories mmon exchange formats CAMP-DX format for mass spectrometry etCDF format for hyphenated data (LC-MS, GC-MS) mzXML format for (LC-MS and MS/MS) rmats for proteomics mzData (PSI proteomics standard Initiative) mzXML (Seattle Proteome Center, sashimi) New: mzML Ask vendors for multiple export options, proprietary formats are no good Format converters are only temporary solutions 6
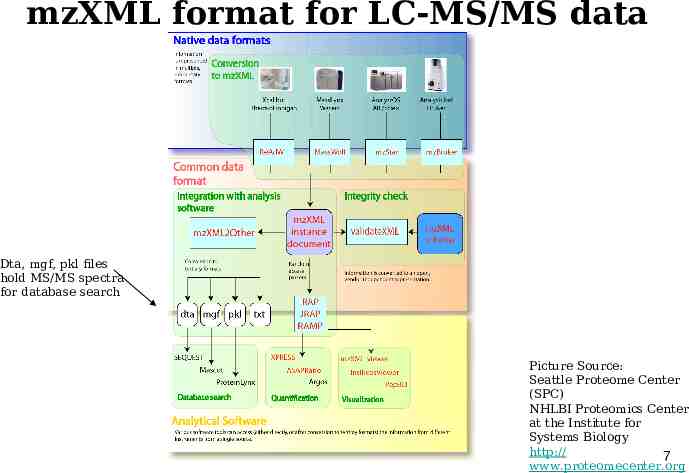
mzXML format for LC-MS/MS data Dta, mgf, pkl files hold MS/MS spectra for database search Picture Source: Seattle Proteome Center (SPC) NHLBI Proteomics Center at the Institute for Systems Biology http:// 7 www.proteomecenter.org
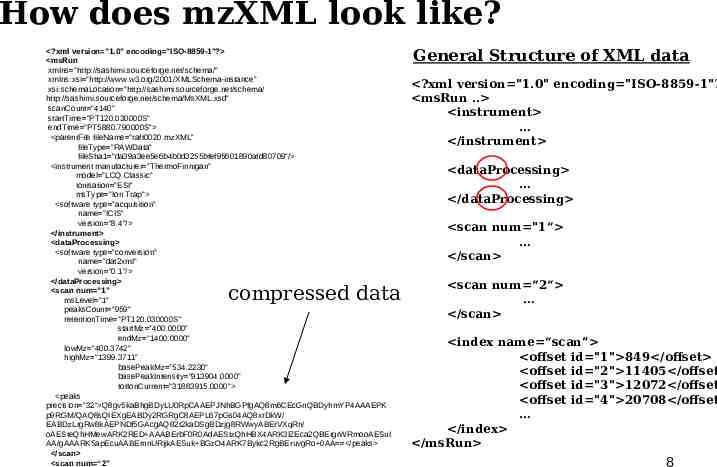
How does mzXML look like? ?xml version "1.0" encoding "ISO-8859-1"? msRun xmlns "http://sashimi.sourceforge.net/schema/" xmlns:xsi "http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation "http://sashimi.sourceforge.net/schema/ http://sashimi.sourceforge.net/schema/MsXML.xsd" scanCount "4140" startTime "PT120.030000S" endTime "PT5880.790000S" parentFile fileName "raft0020.mzXML" fileType "RAWData" fileSha1 "da39a3ee5e6b4b0d3255bfef95601890afd80709"/ instrument manufacturer "ThermoFinnigan" model "LCQ Classic" ionisation "ESI" msType "Ion Trap" software type "acquisition" name "ICIS" version "8.4"/ /instrument dataProcessing software type "conversion" name "dat2xml" version "0.1"/ /dataProcessing scan num "1" msLevel "1" peaksCount "959" retentionTime "PT120.030000S" startMz "400.0000" endMz "1400.0000" lowMz "400.3742" highMz "1399.3711" basePeakMz "534.2230" basePeakIntensity "913904.0000" totIonCurrent "31883915.0000" peaks precision "32" Q8gv5kaBhgBDyLU0RpCAAEPJNhBGPfgAQ8m6CEcGnQBDyhmYP4AAAEPK p9RGM/QAQ8sQIEXgEABDy2RGRgC8AEPL67pGs04AQ8xrDkW/ EABDzLrgRw8kAEPNDf5GAcgAQ82t2kaDSgBDzjg8RWwyABErVXqRn/ oAESteQhHMewARK2RED AAABErbF0R0AdAEStzQhHBX4ARK3lZEca2QBErgrWRmooAESuI AA/gAAARK5apEcuAABErnnURijkAESuk BGzO4ARK7Bykc2RgBEruvgRo 0AA /peaks /scan scan num “2" compressed data General Structure of XML data ?xml version "1.0" encoding "ISO-8859-1"? msRun . instrument /instrument dataProcessing /dataProcessing scan num "1“ /scan scan num “2“ /scan index name “scan” offset id "1" 849 /offset offset id "2" 11405 /offset offset id "3" 12072 /offset offset id "4" 20708 /offset /index /msRun 8
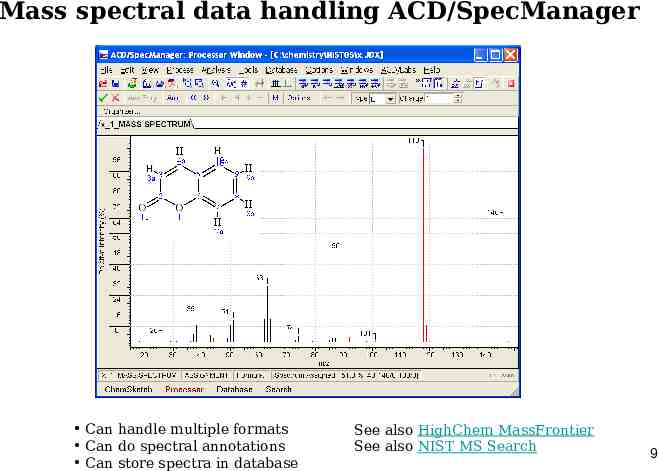
Mass spectral data handling ACD/SpecManager Can handle multiple formats Can do spectral annotations Can store spectra in database See also HighChem MassFrontier See also NIST MS Search 9
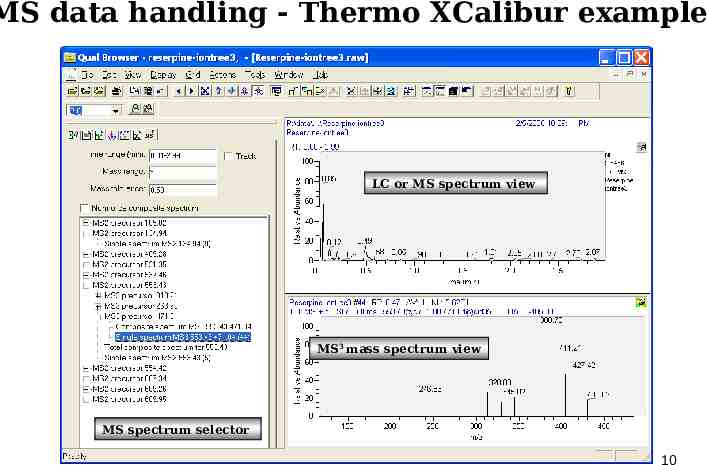
MS data handling - Thermo XCalibur example LC or MS spectrum view MS3 mass spectrum view MS spectrum selector 10
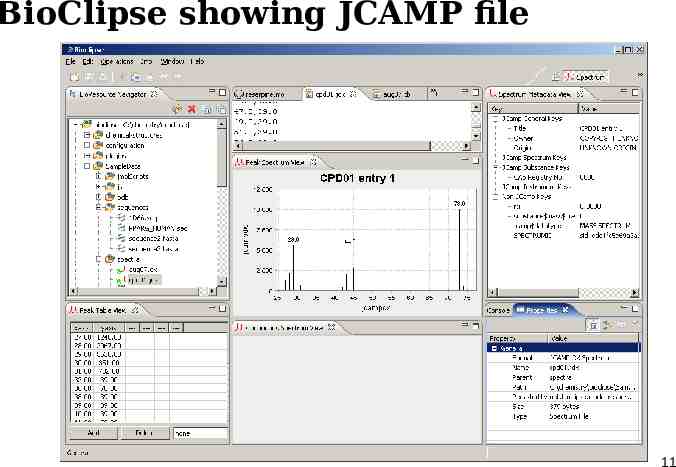
BioClipse showing JCAMP file 11
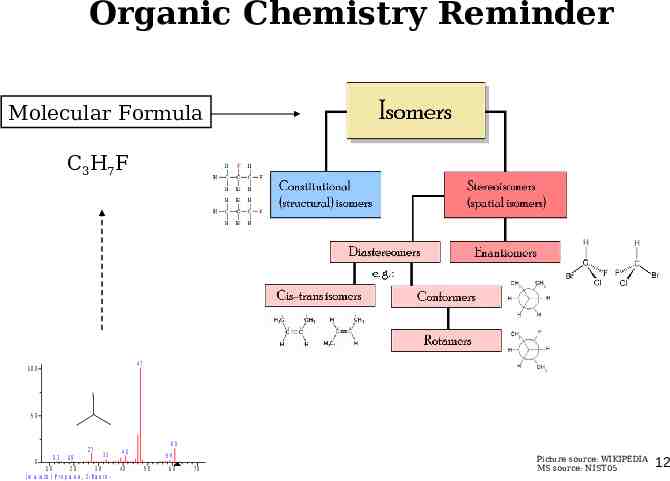
Organic Chemistry Reminder Molecular Formula C3H7F 47 100 F 50 27 0 13 19 33 10 20 30 ( m a in lib ) P r o p a n e , 2 - fl u o r o - 61 41 40 59 50 60 70 Picture source: WIKIPEDIA MS source: NIST05 12
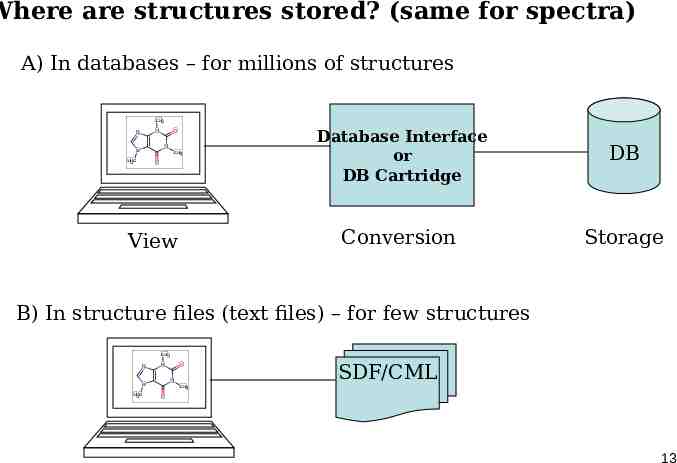
Where are structures stored? (same for spectra) A) In databases – for millions of structures CH3 N N O N N CH3 H3C O View Database Interface or DB Cartridge DB Conversion Storage B) In structure files (text files) – for few structures CH3 N N N N H3C O SDF/CML CH3 O 13
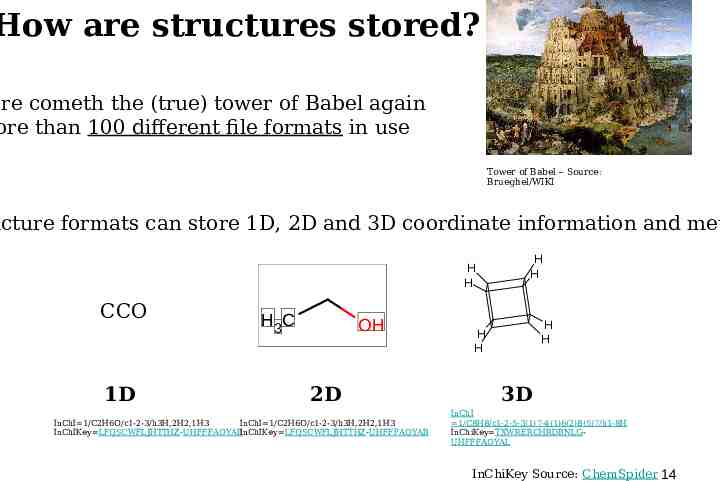
How are structures stored? ere cometh the (true) tower of Babel again ore than 100 different file formats in use Tower of Babel – Source: Brueghel/WIKI ucture formats can store 1D, 2D and 3D coordinate information and met H H CCO 1D H 3C OH 2D InChI 1/C2H6O/c1-2-3/h3H,2H2,1H3 InChI 1/C2H6O/c1-2-3/h3H,2H2,1H3 InChIKey LFQSCWFLJHTTHZ-UHFFFAOYABInChIKey LFQSCWFLJHTTHZ-UHFFFAOYAB H H H H H H 3D InChI 1/C8H8/c1-2-5-3(1)7-4(1)6(2)8(5)7/h1-8H InChiKey TXWRERCHRDBNLGUHFFFAOYAL InChiKey Source: ChemSpider 14

Chemical Structure Handling H3 C H3 C H O H3 C O C C H H3 3 O C H3 C H3 Most common structure formats you need to know:Moronic Acid - CID: 489941 SMILES/SMARTS - Simplified Molecular Input Line Entry Specification SDF/MOL - Structure Data File InChI/InChIkey - IUPAC International Chemical Identifier PDB - Protein Data Bank CML - Chemical Markup Language Some problems: Data format needs to be based on Open Standard (problem with SMILES, ok with CML) Stereo and aromatic bond information needs to be saved (ok with SDF) Format needs to be small in space for millions of compounds (ok with SMILES) SMILES notation needs to be unique (problem with SMILES) 15 Structure representation should be portable and based on Open Standard
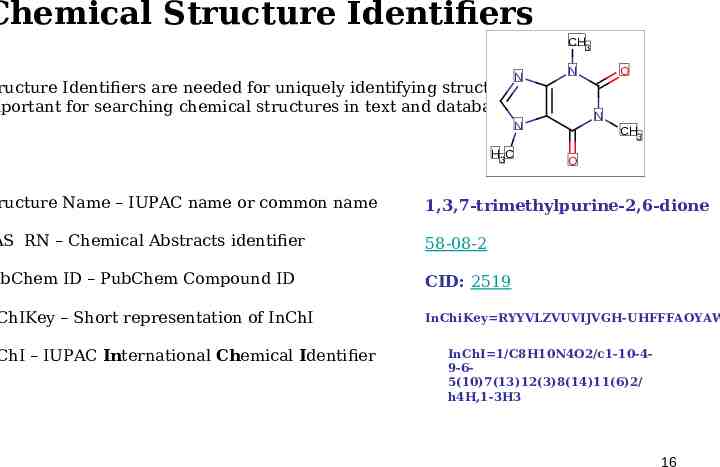
Chemical Structure Identifiers CH3 N ructure Identifiers are needed for uniquely identifying structures mportant for searching chemical structures in text and databases ructure Name – IUPAC name or common name H3C CH3 O 1,3,7-trimethylpurine-2,6-dione 58-08-2 ubChem ID – PubChem Compound ID CID: 2519 ChI – IUPAC International Chemical Identifier O N N AS RN – Chemical Abstracts identifier ChIKey – Short representation of InChI N InChiKey RYYVLZVUVIJVGH-UHFFFAOYAW InChI 1/C8H10N4O2/c1-10-49-65(10)7(13)12(3)8(14)11(6)2/ h4H,1-3H3 16
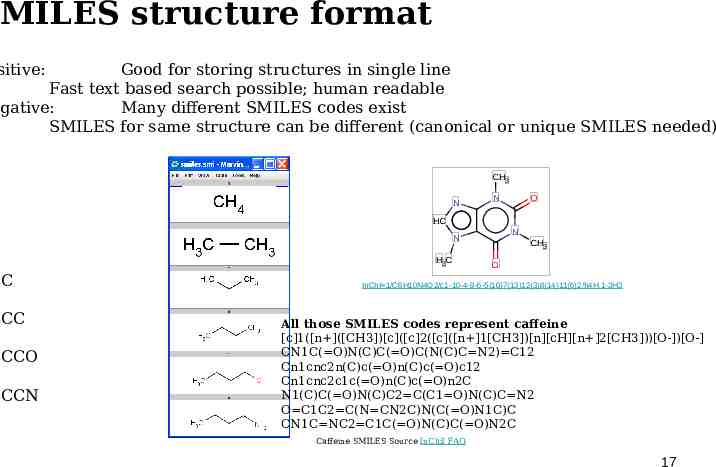
MILES structure format sitive: Good for storing structures in single line Fast text based search possible; human readable gative: Many different SMILES codes exist SMILES for same structure can be different (canonical or unique SMILES needed) CH3 N CCC CCCO CCCN O HC C CC N N N H3C CH3 O InChI 1/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)8(14)11(6)2/h4H,1-3H3 All those SMILES codes represent caffeine [c]1([n ]([CH3])[c]([c]2([c]([n ]1[CH3])[n][cH][n ]2[CH3]))[O-])[O-] CN1C( O)N(C)C( O)C(N(C)C N2) C12 Cn1cnc2n(C)c( O)n(C)c( O)c12 Cn1cnc2c1c( O)n(C)c( O)n2C N1(C)C( O)N(C)C2 C(C1 O)N(C)C N2 O C1C2 C(N CN2C)N(C( O)N1C)C CN1C NC2 C1C( O)N(C)C( O)N2C Caffeine SMILES Source InChiI FAQ 17
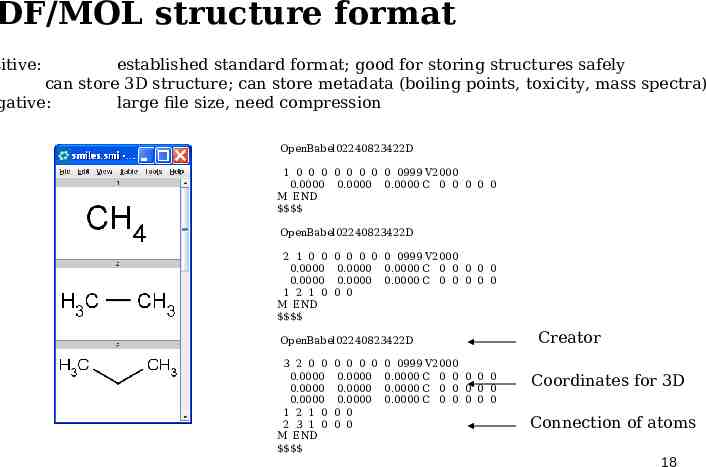
DF/MOL structure format sitive: established standard format; good for storing structures safely can store 3D structure; can store metadata (boiling points, toxicity, mass spectra) gative: large file size, need compression OpenBabel02240823422D 1 0 0 0 0 0 0 0 0 0999 V2000 0.0000 0.0000 0.0000 C 0 0 0 0 0 M END OpenBabel02240823422D 2 1 0 0 0.0000 0.0000 1 2 1 0 M END 0 0 0 0 0 0999 V2000 0.0000 0.0000 C 0 0 0 0 0 0.0000 0.0000 C 0 0 0 0 0 0 0 OpenBabel02240823422D 3 2 0 0 0.0000 0.0000 0.0000 1 2 1 0 2 3 1 0 M END 0 0 0 0 0.0000 0.0000 0.0000 0 0 0 0 0 0999 V2000 0.0000 C 0 0 0 0 0 0.0000 C 0 0 0 0 0 0.0000 C 0 0 0 0 0 Creator Coordinates for 3D Connection of atoms 18
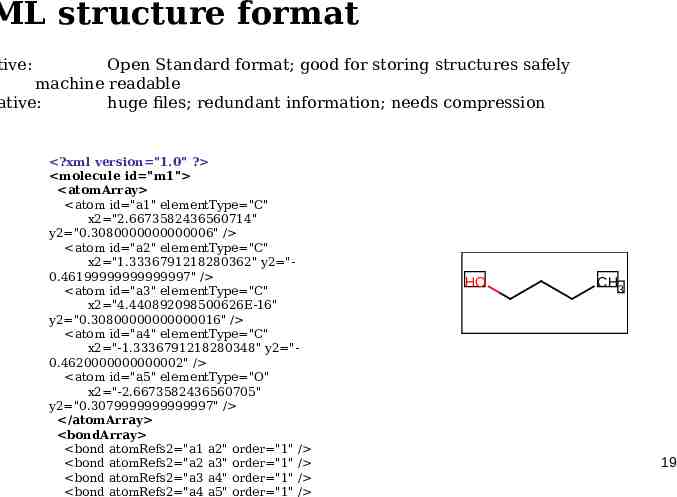
ML structure format tive: Open Standard format; good for storing structures safely machine readable ative: huge files; redundant information; needs compression ?xml version "1.0" ? molecule id "m1" atomArray atom id "a1" elementType "C" x2 "2.6673582436560714" y2 "0.3080000000000006" / atom id "a2" elementType "C" x2 "1.3336791218280362" y2 "0.46199999999999997" / atom id "a3" elementType "C" x2 "4.440892098500626E-16" y2 "0.30800000000000016" / atom id "a4" elementType "C" x2 "-1.3336791218280348" y2 "0.4620000000000002" / atom id "a5" elementType "O" x2 "-2.6673582436560705" y2 "0.3079999999999997" / /atomArray bondArray bond atomRefs2 "a1 a2" order "1" / bond atomRefs2 "a2 a3" order "1" / bond atomRefs2 "a3 a4" order "1" / bond atomRefs2 "a4 a5" order "1" / HO CH3 19
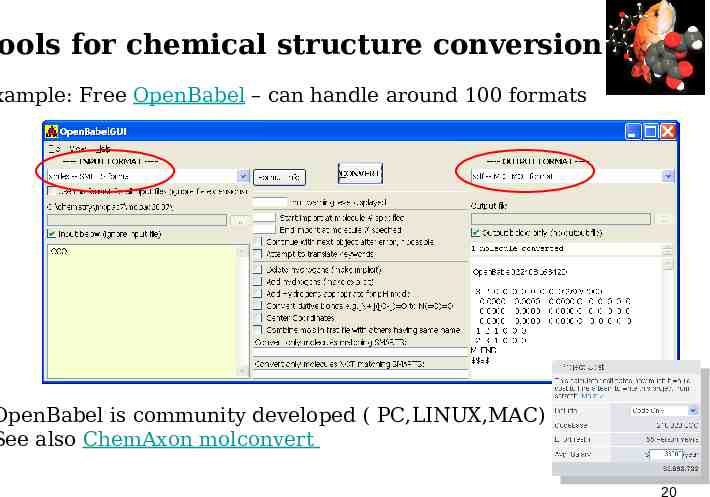
ools for chemical structure conversion xample: Free OpenBabel – can handle around 100 formats OpenBabel is community developed ( PC,LINUX,MAC) See also ChemAxon molconvert 20
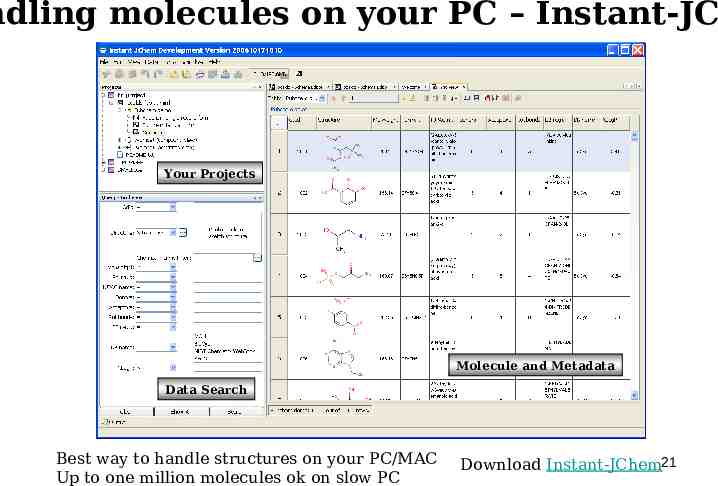
ndling molecules on your PC – Instant-JCh Your Projects Molecule and Metadata Data Search Best way to handle structures on your PC/MAC Up to one million molecules ok on slow PC Download Instant-JChem21

he Last Page - What is important to remember ere are different exchange formats for mass spectral data netCDF, JCAMP, mzXML tadata must be stored together with mass spectra ss spectra should be published in machine readable format (not on pap en Data formats for mass spectral data (in XML) are important ere are different exchange formats for chemical structures SMILES, SDF, MOL, PDB, InChIkey, PDB, CML en Data formats and identifiers for chemical structures are important 22

sks (30 min): stall BioClipse (MAC/PC/LINUX) and open some of the cluded JDX spectra or structures [LINK] stall Instant-JChem (MAC/PC/LINUX) – create a local demo tabase and import the LMSD Structure-data file (SDF) [LIN or diligent students or proteomics PhD candidates: to http://www.ms-utils.org/wiki/pmwiki.php/Main/SoftwareList http://www.proteomecommons.org/tools.jsp http://tools.proteomecenter.org/software.php http://ncrr.pnl.gov/software/ d install one viewer or one visualizer software for MS data. ditionally explain what dta, mgf, pkl files are. 23

5 min): Metabolomics tandards initiative -easing communication and minimizing data loss in a 24
Used Links http://www.bioinformaticssolutions.com/products/peaks/proteinID.php http://geoffhutchison.net/files/BabelTalk04.pdf http://www.google.com/search?hl en&q smiles sdf smarts sdf ppt&btnG Search http://depth-first.com/articles/2007/09/26/pubchem-for-newbies http://depth-first.com/articles/2007/01/24/thirty-two-free-chemistry-databases http://scholar.google.com/scholar?hl en&lr &sa G&oi qs&q cml portable open markup author:h-rzepa http://wwmm.ch.cam.ac.uk/inchifaq/ 25